2XGA
 
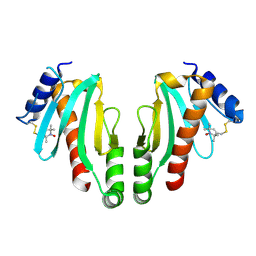 | | MTSL spin-labelled Shigella Flexneri Spa15 | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, SURFACE PRESENTATION OF ANTIGENS PROTEIN SPAK | | Authors: | Lillington, J.E.D, Johnson, S, Lea, S.M. | | Deposit date: | 2010-06-02 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Shigella Flexneri Spa15 Crystal Structure Verified in Solution by Double Electron Electron Resonance.
J.Mol.Biol., 405, 2011
|
|
7P4E
 
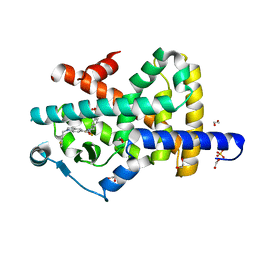 | | Crystal structure of PPARgamma in complex with compound FL217 | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma, SULFATE ION, ... | | Authors: | Ni, X, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7P4K
 
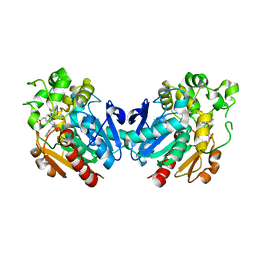 | | Soluble epoxide hydrolase in complex with FL217 | | Descriptor: | Bifunctional epoxide hydrolase 2, ~{N}-[[4-(cyclopropylsulfonylamino)-2-(trifluoromethyl)phenyl]methyl]-1-[(3-fluorophenyl)methyl]indole-5-carboxamide | | Authors: | Ni, X, Kramer, J.S, Lillich, F, Proschak, E, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Dual Partial Peroxisome Proliferator-Activated Receptor gamma Agonists/Soluble Epoxide Hydrolase Inhibitors.
J.Med.Chem., 64, 2021
|
|
2YEQ
 
 | | Structure of PhoD | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALKALINE PHOSPHATASE D, ... | | Authors: | Lillington, J.E.D, Rodriguez, F, Roversi, P, Johnson, S.J, Berks, B, Lea, S.M. | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of the Bacillus Subtilis Phosphodiesterase Phod Reveals an Iron and Calcium-Containing Active Site.
J.Biol.Chem., 289, 2014
|
|
2O3P
 
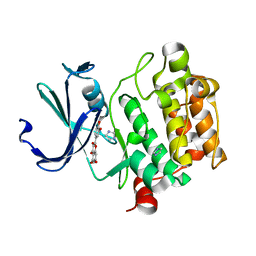 | | Crystal structure of Pim1 with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-01 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
2O65
 
 | | Crystal structure of Pim1 with Pentahydroxyflavone | | Descriptor: | 5,7-DIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
2O64
 
 | | Crystal structure of Pim1 with Quercetagetin | | Descriptor: | 3,5,6,7-TETRAHYDROXY-2-(3,4-DIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
2O63
 
 | | Crystal structure of Pim1 with Myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
4AMF
 
 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
4ALF
 
 | | Pseudomonas fluorescens PhoX in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, ALKALINE PHOSPHATASE PHOX, CALCIUM ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
4A9X
 
 | | Pseudomonas fluorescens PhoX in complex with the substrate analogue AppCp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, MU-OXO-DIIRON, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
4A9V
 
 | | Pseudomonas fluorescens PhoX | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-11-28 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
3ZWU
 
 | | Pseudomonas fluorescens PhoX in complex with vanadate, a transition state analogue | | Descriptor: | ALKALINE PHOSPHATASE PHOX, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yong, S.C, Roversi, P, Lillington, J.E.D, Zeldin, O.B, Garman, E.F, Lea, S.M, Berks, B.C. | | Deposit date: | 2011-08-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Complex Iron-Calcium Cofactor Catalyzing Phosphotransfer Chemistry
Science, 345, 2014
|
|
2W8B
 
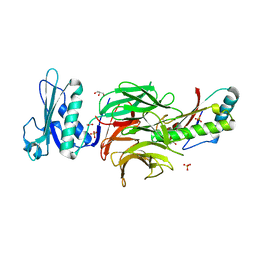 | | Crystal structure of processed TolB in complex with Pal | | Descriptor: | ACETATE ION, GLYCEROL, PEPTIDOGLYCAN-ASSOCIATED LIPOPROTEIN, ... | | Authors: | Sharma, A, Bonsor, D.A, Kleanthous, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-02-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Allosteric Beta-Propeller Signalling in Tolb and its Manipulation by Translocating Colicins.
Embo J., 28, 2009
|
|
5U3B
 
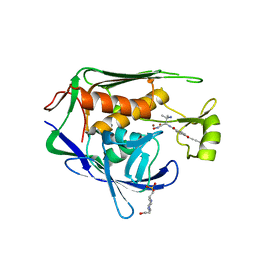 | | Pseudomonas aeruginosa LpxC in complex with NVS-LPXC-01 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(2S)-3-amino-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-[(but-2-yn-1-yl)oxy]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ... | | Authors: | Sprague, E.R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC.
J. Med. Chem., 60, 2017
|
|
5U39
 
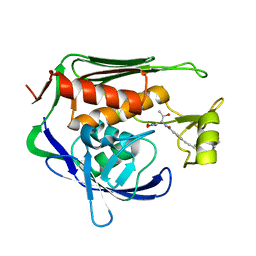 | | Pseudomonas aeruginosa LpxC in complex with CHIR-090 | | Descriptor: | N-{(1S,2R)-2-hydroxy-1-[(hydroxyamino)carbonyl]propyl}-4-{[4-(morpholin-4-ylmethyl)phenyl]ethynyl}benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Sprague, E.R. | | Deposit date: | 2016-12-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC.
J. Med. Chem., 60, 2017
|
|
6CMI
 
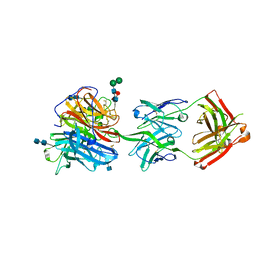 | |
6CMG
 
 | |
7B88
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with S99 inhibitor | | Descriptor: | 3-[5-[3,5-bis(chloranyl)phenyl]-4-phenyl-1,3-oxazol-2-yl]propanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chaikuad, A, Schierle, S, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Oxaprozin Analogues as Selective RXR Agonists with Superior Properties and Pharmacokinetics.
J.Med.Chem., 64, 2021
|
|
7B9O
 
 | | Crystal structure of Retinoic Acid Receptor alpha (RXRA) in complexed with S169 inhibitor | | Descriptor: | 3-(5-(3,5-bis(trifluoromethyl)phenyl)-4-phenyloxazol-2-yl)propanoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Ni, X, Chaikuad, A, Schierle, S, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-14 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Oxaprozin Analogues as Selective RXR Agonists with Superior Properties and Pharmacokinetics.
J.Med.Chem., 64, 2021
|
|
4HQS
 
 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_0659 (Etrx1) from Streptococcus pneumoniae strain TIGR4 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Thioredoxin family protein | | Authors: | Bartual, S.G, Saleh, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-26 | | Release date: | 2013-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Molecular architecture of Streptococcus pneumoniae surface thioredoxin-fold lipoproteins crucial for extracellular oxidative stress resistance and maintenance of virulence.
EMBO Mol Med, 5, 2013
|
|
4HQZ
 
 | | Crystal structure of the pneumoccocal exposed lipoprotein thioredoxin sp_1000 (Etrx2) from Streptococcus pneumoniae strain TIGR4 in complex with 2-hydroxyethyl disulfide | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, DI(HYDROXYETHYL)ETHER, Thioredoxin family protein | | Authors: | Bartual, S.G, Saleh, M, Abdullah, M.R, Jensch, I, Asmat, T.M, Petruschka, L, Pribyl, T, Hermoso, J.A, Hammerschmidt, S. | | Deposit date: | 2012-10-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Molecular architecture of Streptococcus pneumoniae surface thioredoxin-fold lipoproteins crucial for extracellular oxidative stress resistance and maintenance of virulence.
EMBO Mol Med, 5, 2013
|
|
5MWA
 
 | | human sEH Phosphatase in complex with 3-4-3,4-dichlorophenyl-5-phenyl-1,3-oxazol-2-yl-benzoic-acid | | Descriptor: | 3-[4-(3,4-dichlorophenyl)-5-phenyl-1,3-oxazol-2-yl]benzoic acid, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Sorrell, F.J, Fox, N, Chaikuad, A, Knapp, S, Proschak, E. | | Deposit date: | 2017-01-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of first in vivo active inhibitors of soluble epoxide hydrolase (sEH) phosphatase domain.
J.Med.Chem., 2019
|
|
5OH0
 
 | | The Cryo-Electron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M.K, Costa, T.R.D, Redzej, A, Waksman, G. | | Deposit date: | 2017-07-13 | | Release date: | 2017-11-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod.
Structure, 25, 2017
|
|
8GHX
 
 | | Crystal Structure of CelD Cellulase from the Anaerobic Fungus Piromyces finnis | | Descriptor: | 1,2-ETHANEDIOL, Cellulase CelD | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
