5AP9
 
 | | Controlled lid-opening in Thermomyces lanuginosus lipase - a switch for activity and binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Skjold-Joergensen, J, Vind, J, Moroz, O.V, Blagova, E.V, Bhatia, V.K, Svendsen, A, Wilson, K.S, Bjerrum, M.J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controlled lid-opening in Thermomyces lanuginosus lipase- An engineered switch for studying lipase function.
Biochim. Biophys. Acta, 1865, 2017
|
|
6YHT
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | CITRIC ACID, Conk-C1, SULFATE ION | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Molecular Lid Mechanism of K + Channel Blocker Action Revealed by a Cone Peptide.
J.Mol.Biol., 433, 2021
|
|
6YHY
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | Conk-S1 | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Molecular Lid Mechanism of K + Channel Blocker Action Revealed by a Cone Peptide.
J.Mol.Biol., 433, 2021
|
|
6U9G
 
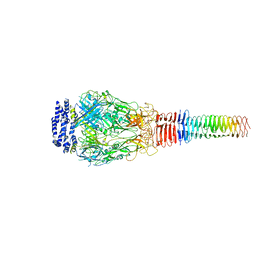 | | Structure of Francisella PdpA-VgrG Complex, half-lidded | | Descriptor: | PdpA, VgrG | | Authors: | Yang, X, Clemens, D.L, Lee, B.-Y, Cui, Y, Zhou, Z.H, Horwitz, M.A. | | Deposit date: | 2019-09-08 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Atomic Structure of the Francisella T6SS Central Spike Reveals a Unique alpha-Helical Lid and a Putative Cargo.
Structure, 27, 2019
|
|
6U9F
 
 | | Structure of Francisella PdpA-VgrG Complex, Lidded | | Descriptor: | PdpA, VgrG | | Authors: | Yang, X, Clemens, D.L, Lee, B.-Y, Cui, Y.X, Horwitz, M.A, Zhou, Z.H. | | Deposit date: | 2019-09-08 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Atomic Structure of the Francisella T6SS Central Spike Reveals a Unique alpha-Helical Lid and a Putative Cargo.
Structure, 27, 2019
|
|
3JCK
 
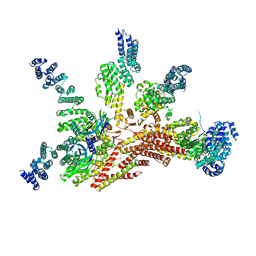 | | Structure of the yeast 26S proteasome lid sub-complex | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN12, 26S proteasome regulatory subunit RPN3, ... | | Authors: | Herzik Jr, M.A, Dambacher, C.M, Worden, E.J, Martin, A, Lander, G.C. | | Deposit date: | 2015-12-20 | | Release date: | 2016-01-20 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structure of the 26S proteasome lid reveals the mechanism of deubiquitinase inhibition.
Elife, 5, 2016
|
|
2LXD
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for LMO2(LIM2)-Ldb1(LID) | | Descriptor: | Rhombotin-2,LIM domain-binding protein 1, ZINC ION | | Authors: | Dastmalchi, S, Wilkinson-White, L, Kwan, A.H, Gamsjaeger, R, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tethered Lmo2(LIM2) /Ldb1(LID) complex.
Protein Sci., 21, 2012
|
|
2MIQ
 
 | | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 of Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J | | Descriptor: | Lysine-specific demethylase lid, ZINC ION | | Authors: | Xu, X, Eletsky, A, Shastry, R, Maglaqui, M, Janjua, H, Xiao, R, Everett, J.K, Sukumaran, D.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of PHD Type 1 Zinc Finger Domain 1 from Lysine-specific Demethylase Lid from Drosophila melanogaster, Northeast Structural Genomics Consortium (NESG) Target FR824J
To be Published
|
|
2MR3
 
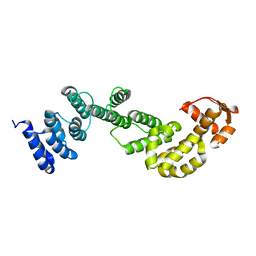 | | A subunit of 26S proteasome lid complex | | Descriptor: | 26S proteasome regulatory subunit RPN9 | | Authors: | Wu, Y, Hu, Y, Jin, C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-02-04 | | Last modified: | 2017-04-12 | | Method: | SOLUTION NMR | | Cite: | Solution structure of yeast Rpn9: insights into proteasome lid assembly.
J. Biol. Chem., 290, 2015
|
|
3J47
 
 | | Formation of an intricate helical bundle dictates the assembly of the 26S proteasome lid | | Descriptor: | 26S proteasome regulatory subunit RPN11, 26S proteasome regulatory subunit RPN12, 26S proteasome regulatory subunit RPN3, ... | | Authors: | Estrin, E, Lopez-Blanco, J.R, Chacon, P, Martin, A. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Formation of an Intricate Helical Bundle Dictates the Assembly of the 26S Proteasome Lid.
Structure, 21, 2013
|
|
3MOE
 
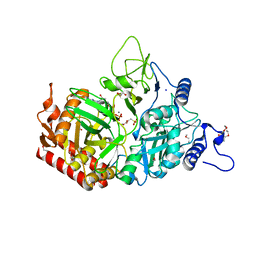 | |
3MOF
 
 | |
3MOH
 
 | |
8GOL
 
 | | crystal structure of SulE | | Descriptor: | 2-[(4-chloranyl-6-methoxy-pyrimidin-2-yl)carbamoylsulfamoyl]benzoic acid, Alpha/beta fold hydrolase, GLYCEROL | | Authors: | Liu, B, Ran, T, Wang, W, He, J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8GOY
 
 | | SulE P44R | | Descriptor: | 5-[(4,6-dimethoxypyrimidin-2-yl)carbamoylsulfamoyl]-1-methyl-pyrazole-4-carboxylic acid, Alpha/beta fold hydrolase, GLYCEROL | | Authors: | Liu, B, He, J, Ran, T, Wang, W. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8GP0
 
 | | crystal structure of SulE | | Descriptor: | Alpha/beta fold hydrolase, CITRIC ACID, GLYCEROL | | Authors: | Liu, B, Ran, T, wang, W, He, J. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7D0N
 
 | | Crystal structure of mouse CRY2 apo form | | Descriptor: | Cryptochrome-2 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-09-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D0M
 
 | | Crystal structure of mouse CRY1 with bound cryoprotectant | | Descriptor: | Cryptochrome-1, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-09-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DLI
 
 | | Crystal structure of mouse CRY1 in complex with KL001 compound | | Descriptor: | 1,2-ETHANEDIOL, Cryptochrome-1, N-[(2R)-3-carbazol-9-yl-2-oxidanyl-propyl]-N-(furan-2-ylmethyl)methanesulfonamide | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-11-27 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7EJ9
 
 | | Alternative crystal structure of mouse Cryptochrome 2 in complex with TH301 compound | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(4-methoxyphenyl)-5,5-bis(oxidanylidene)-4,6-dihydrothieno[3,4-c]pyrazol-3-yl]cyclopentane-1-carboxamide, Cryptochrome-2 | | Authors: | Miller, S.A, Hirota, T. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8SBM
 
 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | Descriptor: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | Authors: | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | Deposit date: | 2023-04-03 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
2OXE
 
 | | Structure of the Human Pancreatic Lipase-related Protein 2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic lipase-related protein 2, ... | | Authors: | Walker, J.R, Davis, T, Seitova, A, Finerty Jr, P.J, Butler-Cole, C, Kozieradzki, I, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-20 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
2PVS
 
 | | Structure of human pancreatic lipase related protein 2 mutant N336Q | | Descriptor: | CALCIUM ION, Pancreatic lipase-related protein 2, SULFATE ION | | Authors: | Spinelli, S, Eydoux, C, Carriere, F, Cambillau, C. | | Deposit date: | 2007-05-10 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human pancreatic lipase-related protein 2 with the lid in an open conformation.
Biochemistry, 47, 2008
|
|
8GUU
 
 | |
8GR6
 
 | | Crystal Structure of pilus-specific Sortase C from Streptococcus sanguinis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Sortase-like protein, ... | | Authors: | Yadav, S, Parijat, P, Krishnan, V. | | Deposit date: | 2022-09-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the pilus-specific sortase from early colonizing oral Streptococcus sanguinis captures an active open-lid conformation.
Int.J.Biol.Macromol., 243, 2023
|
|
