4RL4
 
 | |
7A73
 
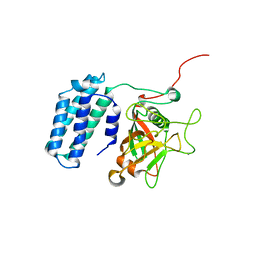 | |
4OOH
 
 | |
7W58
 
 | | Crystal structure of acyl-carrier protein synthase from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, 4'-phosphopantetheinyl transferase, NICKEL (II) ION | | Authors: | Yadav, S, Bhatia, I, Biswal, B.K. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification, structure determination and analysis of Mycobacterium smegmatis acyl-carrier protein synthase (AcpS) crystallized serendipitously.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
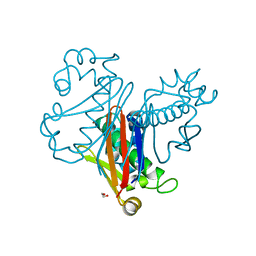
8GUU
 
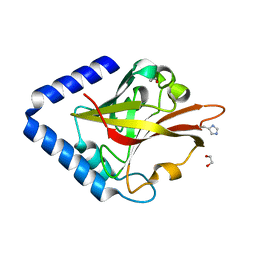 | |
8GR6
 
 | | Crystal Structure of pilus-specific Sortase C from Streptococcus sanguinis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Sortase-like protein, ... | | Authors: | Yadav, S, Parijat, P, Krishnan, V. | | Deposit date: | 2022-09-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the pilus-specific sortase from early colonizing oral Streptococcus sanguinis captures an active open-lid conformation.
Int.J.Biol.Macromol., 243, 2023
|
|
7YSA
 
 | |
7YSY
 
 | |
7YST
 
 | |
7YSM
 
 | |
7YS8
 
 | |
7YS9
 
 | |
7YT0
 
 | |
1BIY
 
 | | STRUCTURE OF DIFERRIC BUFFALO LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Karthikeyan, S, Yadav, S, Singh, T.P. | | Deposit date: | 1998-06-21 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Structure of buffalo lactoferrin at 3.3 A resolution at 277 K.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3ES6
 
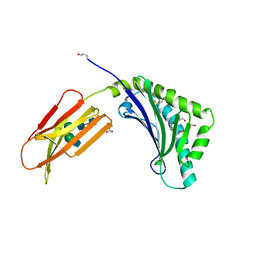 | | Crystal structure of the novel complex formed between Zinc 2-glycoprotein (ZAG) and Prolactin inducible protein (PIP) from human seminal plasma | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, HEXAETHYLENE GLYCOL, ... | | Authors: | Hassan, M.I, Bilgrami, S, Kumar, V, Singh, N, Yadav, S, Kaur, P, Singh, T.P. | | Deposit date: | 2008-10-04 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Crystal structure of the novel complex formed between zinc alpha2-glycoprotein (ZAG) and prolactin-inducible protein (PIP) from human seminal plasma
J.Mol.Biol., 384, 2008
|
|
4Q9E
 
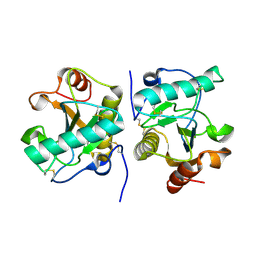 | | Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, GLYCEROL, ... | | Authors: | Yamini, S, Sharma, P, Yadav, S.P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2014-05-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of the ternary complex of peptidoglycan recognition protein, PGRP-S with N-acetyl glucosamine and paranitro benzaldehyde at 2.3 A resolution
to be published
|
|
8H17
 
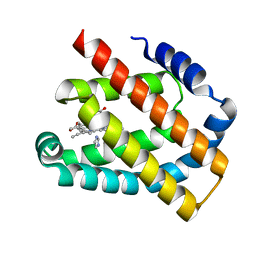 | | Crystal structure of the Globin domain of Thermosynechococcus elongatus BP-1 | | Descriptor: | IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, Tlr1989 protein | | Authors: | Mathur, S, Yadav, S.K, Pal, K.R, Kundu, S. | | Deposit date: | 2022-09-30 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A novel single sensor hemoglobin domain from the thermophilic cyanobacteria Thermosynechococcus elongatus BP-1 exhibits higher pH but lower thermal stability compared to globins from mesophilic organisms.
Int.J.Biol.Macromol., 240, 2023
|
|
3MWN
 
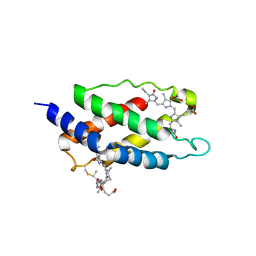 | | Structure of the Novel 14 kDa Fragment of alpha-Subunit of Phycoerythrin from the Starving Cyanobacterium Phormidium Tenue | | Descriptor: | PHYCOCYANOBILIN, PHYCOERYTHRIN | | Authors: | Soni, B.R, Hasan, M.I, Parmar, A, Ethayathulla, A.S, Kumar, R.P, Singh, N.K, Sinha, M, Kaur, P, Yadav, S, Sharma, S, Madamwar, D, Singh, T.P. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the novel 14kDa fragment of alpha-subunit of phycoerythrin from the starving cyanobacterium Phormidium tenue.
J.Struct.Biol., 171, 2010
|
|
7NWG
 
 | | Mammalian pre-termination 80S ribosome with Hybrid P/E- and A/P-site tRNA's bound by Blasticidin S. | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
7NWH
 
 | | Mammalian pre-termination 80S ribosome with eRF1 and eRF3 bound by Blasticidin S. | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
7NWI
 
 | | Mammalian pre-termination 80S ribosome with Empty-A site bound by Blasticidin S | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA+BlaS, 40S ribosomal protein S11, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
6ZB5
 
 | | SARS CoV-2 Spike protein, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
6ZB4
 
 | | SARS CoV-2 Spike protein, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
4GF9
 
 | | Structural insights into the dual strategy of recognition of peptidoglycan recognition protein, PGRP-S: ternary complex of PGRP-S with LPS and fatty acid | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Yadav, S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-03 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the dual strategy of recognition by peptidoglycan recognition protein, PGRP-S: structure of the ternary complex of PGRP-S with lipopolysaccharide and stearic acid.
Plos One, 8, 2013
|
|
7ODL
 
 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-29 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
