2EBI
 
 | | Arabidopsis GT-1 DNA-binding domain with T133D phosphomimetic mutation | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Noto, K, Niyada, E, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2007-02-08 | | Release date: | 2008-02-19 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
2JMW
 
 | | Structure of DNA-Binding Domain of Arabidopsis GT-1 | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Niyada, E, Noto, K, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2006-12-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
1VTT
 
 | | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*G)-3') | | Authors: | Ho, P.S, Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG)
Embo J., 4, 1985
|
|
3OTK
 
 | | Structure and mechanisim of core 2 beta1,6-n-acetylglucosaminyltransferase: a Metal-ion independent gt-a glycosyltransferase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase, ... | | Authors: | Pak, J.E, Rini, J.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic characterization of leukocyte-type core 2 beta 1,6-N-acetylglucosaminyltransferase: a metal-ion-independent GT-A glycosyltransferase.
J.Mol.Biol., 414, 2011
|
|
6WMN
 
 | |
6WMO
 
 | |
6WMM
 
 | |
1H7Q
 
 | | dTDP-MANGANESE COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, ... | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
1H7L
 
 | | dTDP-MAGNESIUM COMPLEX OF SPSA FROM BACILLUS SUBTILIS | | Descriptor: | MAGNESIUM ION, SPORE COAT POLYSACCHARIDE BIOSYNTHESIS PROTEIN SPSA, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Tarbouriech, N, Charnock, S.J, Davies, G.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Three-Dimensional Structures of the Mn and Mg Dtdp Complexes of the Family Gt-2 Glycosyltransferase Spsa: A Comparison with Related Ndp-Sugar Glycosyltransferases.
J.Mol.Biol., 314, 2001
|
|
1XV5
 
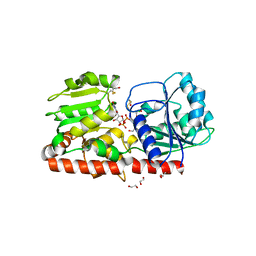 | | alpha-glucosyltransferase (AGT) in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA alpha-glucosyltransferase, ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-10-27 | | Release date: | 2005-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
1YA6
 
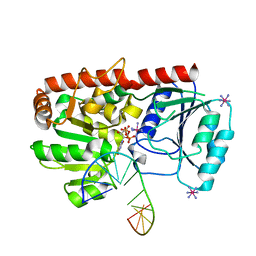 | | alpha-glucosyltransferase in complex with UDP and a 13-mer DNA containing a central A:G mismatch | | Descriptor: | 5'-D(*AP*TP*AP*CP*TP*AP*AP*GP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*T)-3', COBALT HEXAMMINE(III), ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-12-17 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
1Y6G
 
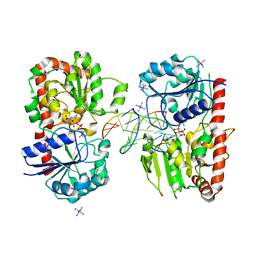 | | alpha-glucosyltransferase in complex with UDP and a 13_mer DNA containing a HMU base at 2.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*T)-3', 5'-D(*GP*AP*TP*AP*CP*TP*(5HU)P*AP*GP*AP*TP*AP*G)-3', ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-12-06 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
1Y6F
 
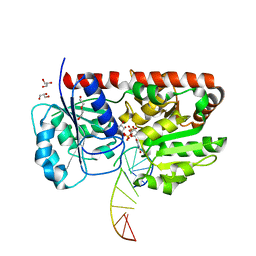 | | alpha-glucosyltransferase in complex with UDP-glucose and DNA containing an abasic site | | Descriptor: | 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*TP*C)-3', 5'-D(*GP*AP*TP*AP*CP*TP*(3DR)P*AP*GP*AP*TP*AP*G)-3', DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-12-06 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
1Y8Z
 
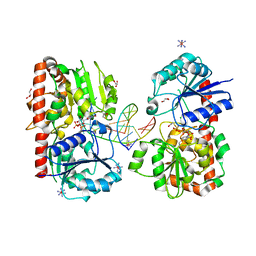 | | alpha-glucosyltransferase in complex with UDP and a 13-mer DNA containing a HMU base at 1.9 A resolution | | Descriptor: | 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*G)-3', 5'-D(*GP*AP*TP*AP*CP*TP*(5HU)P*AP*GP*AP*TP*AP*G)-3', CHLORIDE ION, ... | | Authors: | Lariviere, L, Sommer, N, Morera, S. | | Deposit date: | 2004-12-14 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence of a passive base-flipping mechanism for AGT, an unusual GT-B glycosyltransferase.
J.Mol.Biol., 352, 2005
|
|
2LGT
 
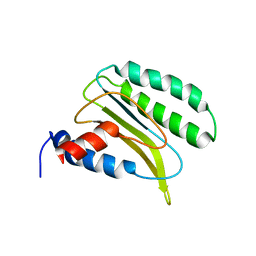 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for QFM(Y)F | | Descriptor: | Eukaryotic peptide chain release factor subunit 1 | | Authors: | Wong, L.E, Li, Y, Pillay, S, Pervushin, K. | | Deposit date: | 2011-08-02 | | Release date: | 2012-03-14 | | Method: | SOLUTION NMR | | Cite: | Selectivity of stop codon recognition in translation termination is modulated by multiple conformations of GTS loop in eRF1
Nucleic Acids Res., 2012
|
|
2C0N
 
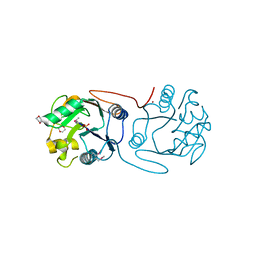 | | Crystal Structure of A197 from STIV | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, A197, NICKEL (II) ION, ... | | Authors: | Larson, E.T, Reiter, D, Young, M, Lawrence, C.M. | | Deposit date: | 2005-09-06 | | Release date: | 2005-09-29 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of A197 from Sulfolobus Turreted Icosahedral Virus: A Crenarchaeal Viral Glycosyltransferase Exhibiting the Gt-A Fold.
J.Virol., 80, 2006
|
|
5KOR
 
 | | Arabidopsis thaliana fucosyltransferase 1 (FUT1) in complex with GDP and a xylo-oligossacharide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rocha, J, de Sanctis, D, Breton, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation.
Plant Cell, 28, 2016
|
|
5KOP
 
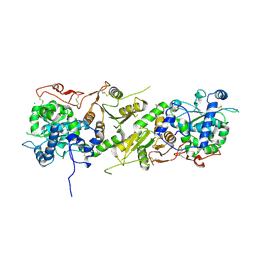 | | Arabidopsis thaliana fucosyltransferase 1 (FUT1) in its apo-form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Galactoside 2-alpha-L-fucosyltransferase | | Authors: | Rocha, J, de Sanctis, D, Breton, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-10-12 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Arabidopsis thaliana FUT1 Reveals a Variant of the GT-B Class Fold and Provides Insight into Xyloglucan Fucosylation.
Plant Cell, 28, 2016
|
|
1GOT
 
 | | HETEROTRIMERIC COMPLEX OF A GT-ALPHA/GI-ALPHA CHIMERA AND THE GT-BETA-GAMMA SUBUNITS | | Descriptor: | GT-ALPHA/GI-ALPHA CHIMERA, GT-BETA, GT-GAMMA, ... | | Authors: | Lambright, D.G, Sondek, J, Bohm, A, Skiba, N.P, Hamm, H.E, Sigler, P.B. | | Deposit date: | 1996-08-07 | | Release date: | 1997-03-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure of a heterotrimeric G protein.
Nature, 379, 1996
|
|
1FQK
 
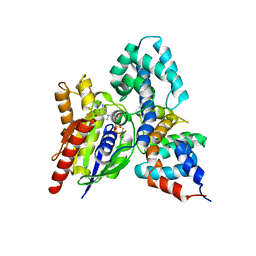 | | CRYSTAL STRUCTURE OF THE HETERODIMERIC COMPLEX OF THE RGS DOMAIN OF RGS9, AND THE GT/I1 CHIMERA ALPHA SUBUNIT [(RGS9)-(GT/I1ALPHA)-(GDP)-(ALF4-)-(MG2+)] | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(t) subunit alpha-1,Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(t) subunit alpha-1, MAGNESIUM ION, ... | | Authors: | Slep, K.C, Kercher, M.A, He, W, Cowan, C.W, Wensel, T.G, Sigler, P.B. | | Deposit date: | 2000-09-05 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A.
Nature, 409, 2001
|
|
1FQJ
 
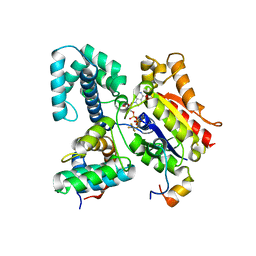 | | CRYSTAL STRUCTURE OF THE HETEROTRIMERIC COMPLEX OF THE RGS DOMAIN OF RGS9, THE GAMMA SUBUNIT OF PHOSPHODIESTERASE AND THE GT/I1 CHIMERA ALPHA SUBUNIT [(RGS9)-(PDEGAMMA)-(GT/I1ALPHA)-(GDP)-(ALF4-)-(MG2+)] | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(t) subunit alpha-1,Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(t) subunit alpha-1, MAGNESIUM ION, ... | | Authors: | Slep, K.C, Kercher, M.A, He, W, Cowan, C.W, Wensel, T.G, Sigler, P.B. | | Deposit date: | 2000-09-05 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural determinants for regulation of phosphodiesterase by a G protein at 2.0 A.
Nature, 409, 2001
|
|
4R9V
 
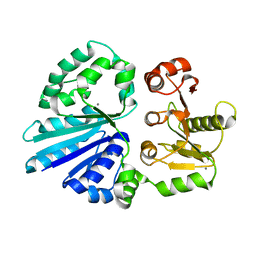 | | Crystal structure of sialyltransferase from photobacterium damselae, residues 113-497 corresponding to the gt-b domain | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Li, Y, Huynh, N, Chen, X, Fisher, A.J. | | Deposit date: | 2014-09-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4DMW
 
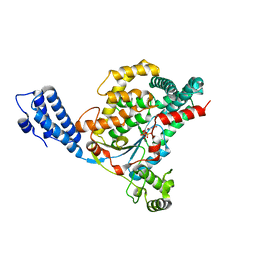 | | Crystal structure of the GT domain of Clostridium difficile toxin A (TcdA) in complex with UDP and Manganese | | Descriptor: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Scarselli, M, Maione, D, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
4DMV
 
 | | Crystal structure of the GT domain of Clostridium difficile Toxin A | | Descriptor: | Toxin A | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Maione, D, Scarselli, M, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
7L1E
 
 | | The Crystal Structure of Bromide-Bound GtACR1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Anion channelrhodopsin-1, BROMIDE ION, ... | | Authors: | Li, H, Huang, C.Y, Wang, M, Spudich, J.L, Zheng, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The crystal structure of bromide-bound Gt ACR1 reveals a pre-activated state in the transmembrane anion tunnel.
Elife, 10, 2021
|
|
