5AAR
 
 | |
6TLH
 
 | |
2DZO
 
 | |
2DZN
 
 | |
1N11
 
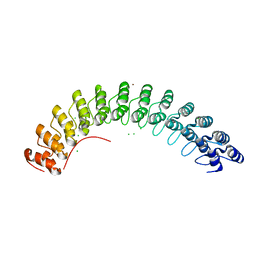 | | D34 REGION OF HUMAN ANKYRIN-R AND LINKER | | Descriptor: | Ankyrin, BROMIDE ION, CHLORIDE ION | | Authors: | Michaely, P, Tomchick, D.R, Machius, M, Anderson, R.G.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a 12 ANK repeat stack from human ankyrinR
Embo J., 21, 2002
|
|
4G8K
 
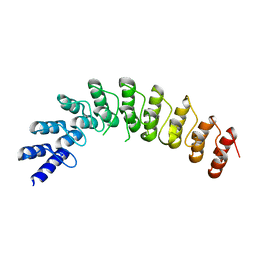 | |
4G8L
 
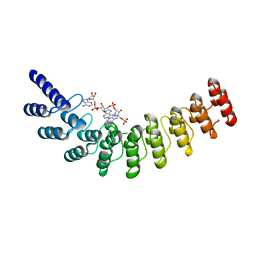 | | Active state of intact sensor domain of human RNase L with 2-5A bound | | Descriptor: | 2-5A-dependent ribonuclease, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE | | Authors: | Han, Y, Whitney, G, Donovan, J, Korennykh, A. | | Deposit date: | 2012-07-23 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Innate Immune Messenger 2-5A Tethers Human RNase L into Active High-Order Complexes.
Cell Rep, 2, 2012
|
|
4RLV
 
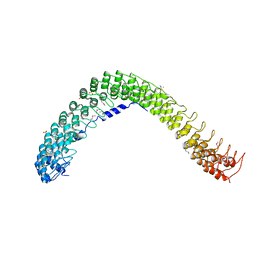 | |
4RLY
 
 | |
3C5R
 
 | |
2NYJ
 
 | | Crystal structure of the ankyrin repeat domain of TRPV1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Ankyrin Repeats of TRPV1 Bind Multiple Ligands and Modulate Channel Sensitivity.
Neuron, 54, 2007
|
|
3DEO
 
 | |
3DEP
 
 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
2PNN
 
 | | Crystal Structure of the Ankyrin Repeat Domain of Trpv1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily V member 1 | | Authors: | Jin, X, Gaudet, R. | | Deposit date: | 2007-04-24 | | Release date: | 2007-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Ankyrin Repeats of TRPV1 Bind Multiple Ligands and Modulate Channel Sensitivity.
Neuron, 54, 2007
|
|
4DX1
 
 | | Crystal structure of the human TRPV4 ankyrin repeat domain | | Descriptor: | GLYCEROL, PHOSPHATE ION, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Inada, H, Gaudet, R. | | Deposit date: | 2012-02-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel.
Biochemistry, 51, 2012
|
|
4DX2
 
 | | Crystal structure of the human TRPV4 ankyrin repeat domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Transient receptor potential cation channel subfamily V member 4, ... | | Authors: | Inada, H, Gaudet, R. | | Deposit date: | 2012-02-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel.
Biochemistry, 51, 2012
|
|
1SW6
 
 | |
1IXV
 
 | | Crystal Structure Analysis of homolog of oncoprotein gankyrin, an interactor of Rb and CDK4/6 | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Padmanabhan, B, Adachi, N, Kataoka, K, Horikoshi, M. | | Deposit date: | 2002-07-09 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the homolog of the oncoprotein gankyrin, an interactor of Rb and CDK4/6
J.BIOL.CHEM., 279, 2004
|
|
1K1B
 
 | | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family | | Descriptor: | B-cell lymphoma 3-encoded protein | | Authors: | Michel, F, Soler-Lopez, M, Petosa, C, Cramer, P, Siebenlist, U, Mueller, C.W. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family.
EMBO J., 20, 2001
|
|
1K1A
 
 | | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family | | Descriptor: | B-cell lymphoma 3-encoded protein | | Authors: | Michel, F, Soler-Lopez, M, Petosa, C, Cramer, P, Siebenlist, U, Mueller, C.W. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family.
EMBO J., 20, 2001
|
|
1WDY
 
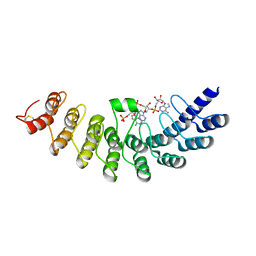 | | Crystal structure of ribonuclease | | Descriptor: | 2-5A-dependent ribonuclease, 5'-O-MONOPHOSPHORYLADENYLYL(2'->5')ADENYLYL(2'->5')ADENOSINE | | Authors: | Tanaka, N, Nakanishi, M, Kusakabe, Y, Goto, Y, Kitade, Y, Nakamura, K.T. | | Deposit date: | 2004-05-19 | | Release date: | 2004-10-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of 2',5'-linked oligoadenylates by human ribonuclease L
Embo J., 23, 2004
|
|
1WG0
 
 | | Structural comparison of Nas6p protein structures in two different crystal forms | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Nakamura, Y, Umehara, T, Tanaka, A, Horikoshi, M, Yokoyama, S, Padmanabhan, B, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2005-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural comparison of Nas6p protein structures in two different crystal forms
To be Published
|
|
4OT9
 
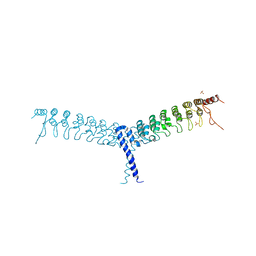 | | crystal structure of the C-terminal domain of p100/NF-kB2 | | Descriptor: | Nuclear factor NF-kappa-B p100 subunit, SULFATE ION | | Authors: | Tao, Z.H, Huang, D.B, Fusco, A, Gupta, K, Ware, C.F, Duynne, G.V. | | Deposit date: | 2014-02-13 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | p100/I kappa B delta sequesters and inhibits NF-kappa B through kappaBsome formation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3JXJ
 
 | |
3JXI
 
 | |
