4ILW
 
 | | Complex of matrix metalloproteinase-10 catalytic domain (MMP-10cd) with tissue inhibitor of metalloproteinases-2 (TIMP-2) | | Descriptor: | CALCIUM ION, Metalloproteinase inhibitor 2, Stromelysin-2, ... | | Authors: | Batra, J, Soares, A.S, Radisky, E.S. | | Deposit date: | 2013-01-01 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Matrix Metalloproteinase-10/TIMP-2 Structure and Analyses Define Conserved Core Interactions and Diverse Exosite Interactions in MMP/TIMP Complexes.
Plos One, 8, 2013
|
|
3V96
 
 | | Complex of matrix metalloproteinase-10 catalytic domain (MMP-10cd) with tissue inhibitor of metalloproteinases-1 (TIMP-1) | | Descriptor: | CALCIUM ION, Metalloproteinase inhibitor 1, PHOSPHATE ION, ... | | Authors: | Batra, J, Soares, A.S, Radisky, E.S. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Matrix metalloproteinase-10 (MMP-10) interaction with tissue inhibitors of metalloproteinases TIMP-1 and TIMP-2: binding studies and crystal structure.
J.Biol.Chem., 287, 2012
|
|
3QLN
 
 | | Crystal structure of ATRX ADD domain in free state | | Descriptor: | Transcriptional regulator ATRX, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QL9
 
 | |
3QLA
 
 | | Hexagonal complex structure of ATRX ADD bound to H3K9me3 peptide | | Descriptor: | POTASSIUM ION, Transcriptional regulator ATRX, ZINC ION, ... | | Authors: | Xiang, B, Li, H. | | Deposit date: | 2011-02-02 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QLC
 
 | |
4W5A
 
 | | Complex structure of ATRX ADD bound to H3K9me3S10ph peptide | | Descriptor: | Peptide from Histone H3.3, Transcriptional regulator ATRX, ZINC ION | | Authors: | Zhao, D, Xiang, B, Li, H. | | Deposit date: | 2014-08-17 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7FBR
 
 | |
7FBV
 
 | |
9L45
 
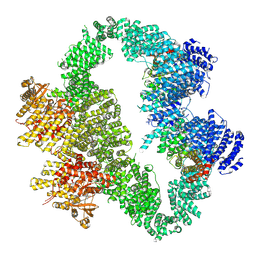 | | ATR-ATRIP bound with VE-822 | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR, VE-822, ... | | Authors: | Wang, G. | | Deposit date: | 2024-12-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 2025
|
|
9L43
 
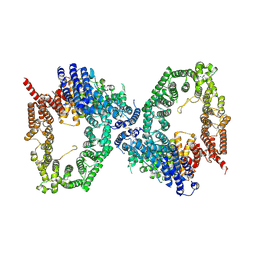 | | ATR Spiral -ATRIP bound with VE-822 | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR, ZINC ION | | Authors: | Wang, G. | | Deposit date: | 2024-12-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 2025
|
|
9L4D
 
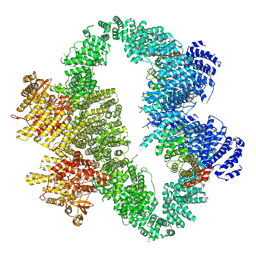 | | ATR-ATRIP bound with RP-3500 | | Descriptor: | ATR-interacting protein, RP-3500, Serine/threonine-protein kinase ATR, ... | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 2025
|
|
9L4C
 
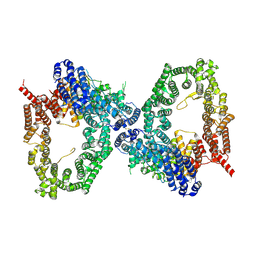 | | ATR Spiral -ATRIP bound with RP-3500 | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR, ZINC ION | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 2025
|
|
9L46
 
 | | ATR-ATRIP-bound with AMP-PNP | | Descriptor: | ATR-interacting protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase ATR | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (6.29 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 2025
|
|
9L4F
 
 | | ATR-ATRIP bound with ATPgammaS | | Descriptor: | ATR-interacting protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase ATR | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (6.22 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 2025
|
|
5YZ0
 
 | | Cryo-EM Structure of human ATR-ATRIP complex | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR | | Authors: | Rao, Q, Liu, M, Tian, Y, Wu, Z, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cryo-EM structure of human ATR-ATRIP complex.
Cell Res., 28, 2018
|
|
8W42
 
 | | X-ray crystal structure of V30M-TTR in complex with resveratrol | | Descriptor: | RESVERATROL, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
4JAZ
 
 | | Crystal structure of the complex between PPARgamma LBD and trans-resveratrol | | Descriptor: | Peroxisome proliferator-activated receptor gamma, RESVERATROL | | Authors: | Pochetti, G, Capelli, D, Montanari, R, Calleri, E, Moaddel, R, Temporini, C. | | Deposit date: | 2013-02-19 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Resveratrol and Its Metabolites Bind to PPARs.
Chembiochem, 15, 2014
|
|
4PP6
 
 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Resveratrol | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, RESVERATROL | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-26 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
1TE7
 
 | | Solution NMR Structure of Protein yqfB from Escherichia coli. Northeast Structural Genomics Consortium Target ET99 | | Descriptor: | Hypothetical UPF0267 protein yqfB | | Authors: | Atreya, H.S, Shen, Y, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-24 | | Release date: | 2005-01-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | G-Matrix Fourier Transform NOESY-Based Protocol for High-Quality Protein Structure Determination
J.Am.Chem.Soc., 127, 2005
|
|
9GO9
 
 | |
9GOA
 
 | |
4IPH
 
 | | Structure of N-terminal domain of RPA70 in complex with VU079104 inhibitor | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, ~{N}-(2,3-dimethylphenyl)-7-oxidanylidene-12-sulfanylidene-5,11-dithia-1,8-diazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,9-triene-10-carboxamide | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IGK
 
 | |
4G1L
 
 | |
