8H77
 
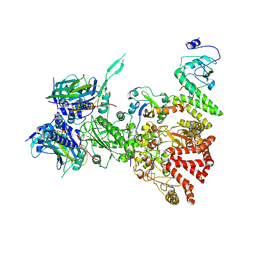 | | Hsp90-AhR-p23-XAP2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7Y04
 
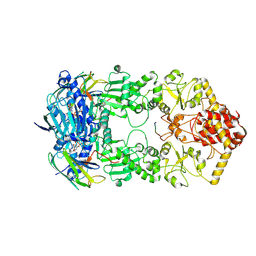 | | Hsp90-AhR-p23 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aryl hydrocarbon receptor, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-06-03 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7ZUB
 
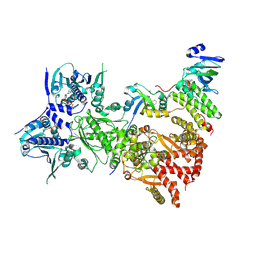 | | Cryo-EM structure of the indirubin-bound Hsp90-XAP2-AHR complex | | Descriptor: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, ... | | Authors: | Gruszczyk, J, Savva, C.G, Lai-Kee-Him, J, Bous, J, Ancelin, A, Kwong, H.S, Grandvuillemin, L, Bourguet, W. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Nat Commun, 13, 2022
|
|
5V0L
 
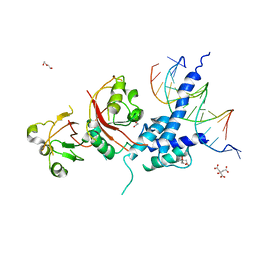 | | Crystal structure of the AHR-ARNT heterodimer in complex with the DRE | | Descriptor: | Aryl hydrocarbon receptor, Aryl hydrocarbon receptor nuclear translocator, CITRIC ACID, ... | | Authors: | Seok, S.-H, Lee, W, Jiang, L, Bradfield, C.A, Xing, Y. | | Deposit date: | 2017-02-28 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural hierarchy controlling dimerization and target DNA recognition in the AHR transcriptional complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y7Y
 
 | | Crystal structure of AhRR/ARNT complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Aryl hydrocarbon receptor repressor, GLYCEROL | | Authors: | Sakurai, S, Shimizu, T, Ohto, U. | | Deposit date: | 2017-08-18 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AhRR-ARNT heterodimer reveals the structural basis of the repression of AhR-mediated transcription.
J. Biol. Chem., 292, 2017
|
|
6OQP
 
 | | U-AITx-Ate1 | | Descriptor: | SER-LYS-TRP-ILE-CYS-ALA-ASN-ARG-SER-VAL-CYS-PRO-ILE | | Authors: | Elnahriry, K.A, Wai, D.C.C, Norton, R.S. | | Deposit date: | 2019-04-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterisation of a novel peptide from the Australian sea anemone Actinia tenebrosa.
Toxicon, 168, 2019
|
|
6O6I
 
 | |
4ZC4
 
 | | Crystal structure of LARP1-unique domain DM15 | | Descriptor: | La-related protein 1, SULFATE ION | | Authors: | Lahr, R.M, Berman, A.J. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5'TOP sequence.
Nucleic Acids Res., 43, 2015
|
|
1Y47
 
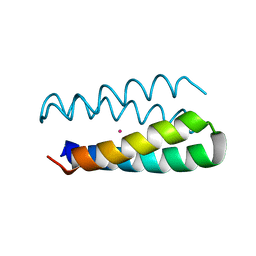 | | Structural studies of designed alpha-helical hairpins | | Descriptor: | CADMIUM ION, dueferri (DF2) | | Authors: | Lahr, S.J, Engel, D.E, Stayrook, S.E, Maglio, O, North, B, Geremia, S, Lombardi, A, DeGrado, W.F. | | Deposit date: | 2004-11-30 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis and design of turns in alpha-helical hairpins
J.Mol.Biol., 346, 2005
|
|
3M66
 
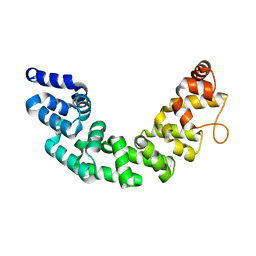 | | Crystal structure of human Mitochondrial Transcription Termination Factor 3 | | Descriptor: | mTERF domain-containing protein 1, mitochondrial | | Authors: | Spahr, H, Samuelsson, T, Hallberg, B.M, Gustafsson, C.M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain.
Biochem.Biophys.Res.Commun., 397, 2010
|
|
1F1E
 
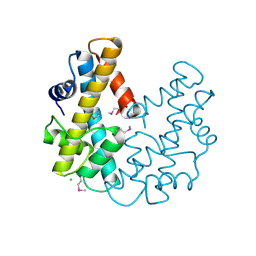 | | CRYSTAL STRUCTURE OF THE HISTONE FROM METHANOPYRUS KANDLERI | | Descriptor: | CHLORIDE ION, HISTONE FOLD PROTEIN | | Authors: | Fahrner, R.L, Cascio, D, Lake, J.A, Slesarev, A. | | Deposit date: | 2000-05-18 | | Release date: | 2001-10-31 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | An ancestral nuclear protein assembly: crystal structure of the Methanopyrus kandleri histone.
Protein Sci., 10, 2001
|
|
1MFT
 
 | | Crystal Structure Of Four-Helix Bundle Model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Lahr, S.J, Stayrook, S.E, North, B, Kaplan, J, Geremia, S, DeGrado, W. | | Deposit date: | 2002-08-13 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
1PG1
 
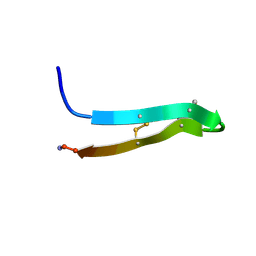 | | PROTEGRIN 1 (PG1) FROM PORCINE LEUKOCYTES, NMR, 20 STRUCTURES | | Descriptor: | PROTEGRIN-1 | | Authors: | Fahrner, R.L, Dieckmann, T, Harwig, S.S.L, Lehrer, R.I, Eisenberg, D, Feigon, J. | | Deposit date: | 1998-03-20 | | Release date: | 1998-05-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes.
Chem.Biol., 3, 1996
|
|
3H0G
 
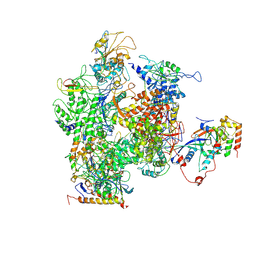 | | RNA Polymerase II from Schizosaccharomyces pombe | | Descriptor: | DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Spahr, H, Calero, G, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2009-04-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Schizosacharomyces pombe RNA polymerase II at 3.6-A resolution.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4FP9
 
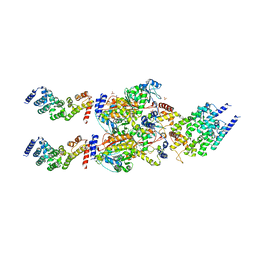 | | Human MTERF4-NSUN4 protein complex | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, mTERF domain-containing protein 2, ... | | Authors: | Spahr, H, Hallberg, B.M. | | Deposit date: | 2012-06-21 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5ORM
 
 | |
5ORQ
 
 | |
5C0V
 
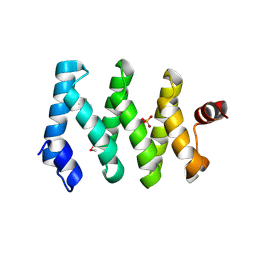 | | Structure of the LARP1-unique domain DM15 | | Descriptor: | La-related protein 1, SULFATE ION | | Authors: | Lahr, R.M, Berman, A.J. | | Deposit date: | 2015-06-12 | | Release date: | 2015-08-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The La-related protein 1-specific domain repurposes HEAT-like repeats to directly bind a 5'TOP sequence.
Nucleic Acids Res., 43, 2015
|
|
8SEM
 
 | |
7VNI
 
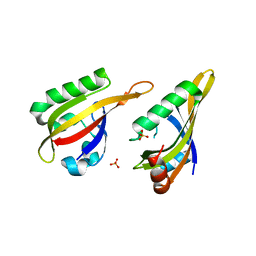 | | AHR-ARNT PAS-B heterodimer | | Descriptor: | Ahr homolog spineless, Aryl hydrocarbon receptor nuclear translocator, SULFATE ION | | Authors: | Dai, S.Y. | | Deposit date: | 2021-10-11 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural insight into the ligand binding mechanism of aryl hydrocarbon receptor.
Nat Commun, 13, 2022
|
|
1F9N
 
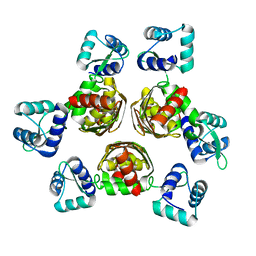 | | CRYSTAL STRUCTURE OF AHRC, THE ARGININE REPRESSOR/ACTIVATOR PROTEIN FROM BACILLUS SUBTILIS | | Descriptor: | ARGININE REPRESSOR/ACTIVATOR PROTEIN | | Authors: | Dennis, C.A, Glykos, N.M, Parsons, M.R, Phillips, S.E.V. | | Deposit date: | 2000-07-11 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6F1E
 
 | | Crystal structure of olive flounder [Paralichthys olivaceus] interferon gamma at 2.3 Angstrom resolution | | Descriptor: | Interferon gamma | | Authors: | Kolenko, P, Kolarova, L, Zahradnik, J, Schneider, B. | | Deposit date: | 2017-11-21 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Interferons type II and their receptors R1 and R2 in fish species: Evolution, structure, and function.
Fish Shellfish Immunol., 79, 2018
|
|
1U7J
 
 | | Solution structure of a diiron protein model | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Maglio, O, Nastri, F, Calhoun, J.R, Lahr, S, Pavone, V, DeGrado, W.F, Lombardi, A. | | Deposit date: | 2004-08-04 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
1U7M
 
 | | Solution structure of a diiron protein model: Due Ferri(II) turn mutant | | Descriptor: | Four-helix bundle model, ZINC ION | | Authors: | Maglio, O, Nastri, F, Calhoun, J.R, Lahr, S, Pavone, V, DeGrado, W.F, Lombardi, A. | | Deposit date: | 2004-08-04 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis and Design of Turns in alpha-Helical Hairpins
J.Mol.Biol., 346, 2005
|
|
6GG1
 
 | | Structure of PROSS-edited human interleukin 24 | | Descriptor: | Interleukin-24, NICKEL (II) ION, SULFATE ION | | Authors: | Kolenko, P, Zahradnik, J, Kolarova, L, Schneider, B. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Flexible regions govern promiscuous binding of IL-24 to receptors IL-20R1 and IL-22R1.
Febs J., 286, 2019
|
|
