4WD9
 
 | |
9BIW
 
 | |
4YAR
 
 | | 2-Hydroxyethylphosphonate dioxygenase (HEPD) E176H | | Descriptor: | 2-hydroxyethylphosphonate dioxygenase, ACETATE ION, CADMIUM ION, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2015-02-17 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Common Late-Stage Intermediate in Catalysis by 2-Hydroxyethyl-phosphonate Dioxygenase and Methylphosphonate Synthase.
J.Am.Chem.Soc., 137, 2015
|
|
6VSK
 
 | | Crystal structure of the P-Rex1 DEP1 domain | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Tesmer, J, Ravala, S.K. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The first DEP domain of the RhoGEF P-Rex1 autoinhibits activity and contributes to membrane binding.
J.Biol.Chem., 295, 2020
|
|
6VL9
 
 | | Anti-PEG antibody 6-3 Fab fragment in complex with PEG | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 6-3 Fab heavy chain, 6-3 Fab light chain | | Authors: | Nicely, N.I, Huckaby, J.T, Lai, S.K, Jacobs, T.M. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure of an anti-PEG antibody reveals an open ring that captures highly flexible PEG polymers
Commun Chem, 3, 2020
|
|
2XEE
 
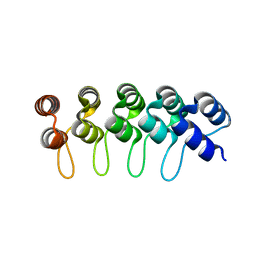 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | NI3C DARPIN MUTANT5 | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-14 | | Release date: | 2010-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
2XEH
 
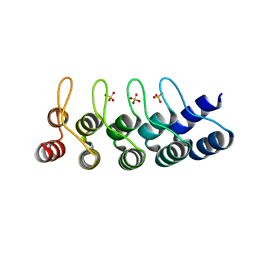 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | NI3C MUT6, SULFATE ION | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-14 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
2XTH
 
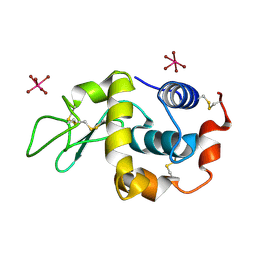 | | K2PtBr6 binding to lysozyme | | Descriptor: | HEXABROMOPLATINATE(IV), LYSOZYME C | | Authors: | Helliwell, J.R, Bell, A.M.T, Bryant, P, Fisher, S, Habash, J, Helliwell, M, Margiolaki, I, Kaenket, S, Watier, Y, Wright, J, Yalamanchili, S.K. | | Deposit date: | 2010-10-07 | | Release date: | 2010-12-08 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-Dependent Analysis of K2Ptbr6 Binding to Lysozyme Studied by Protein Powder and Single Crystal X-Ray Analysis
Z.Kristallogr., 225, 2010
|
|
1QJZ
 
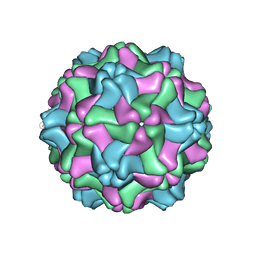 | | Three Dimensional Structure of Physalis Mottle Virus : Implications for the Viral Assembly | | Descriptor: | COAT PROTEIN | | Authors: | Krishna, S.S, Hiremath, C.N, Munshi, S.K, Prahadeeswaran, D, Sastri, M, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 1999-07-07 | | Release date: | 1999-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Three Dimensional Structure of Physalis Movirus: Implications for the Viral Assembly
J.Mol.Biol., 289, 1999
|
|
7AQO
 
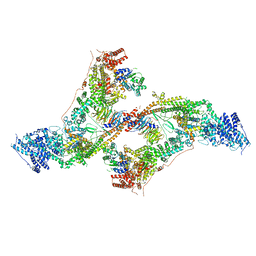 | | yeast THO-Sub2 complex dimer | | Descriptor: | BJ4_G0025130.mRNA.1.CDS.1, EM14S01-3B_G0007820.mRNA.1.CDS.1, TEX1 isoform 1, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-22 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
7APX
 
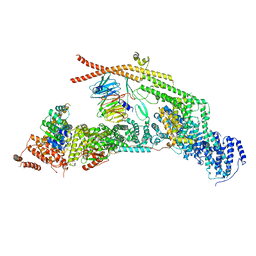 | | yeast THO-Sub2 complex | | Descriptor: | ATP-dependent RNA helicase SUB2, Protein TEX1, THO complex subunit 2,Tho2, ... | | Authors: | Schuller, S.K, Schuller, J.M, Prabu, R.J, Baumgartner, M, Bonneau, F, basquin, J, Conti, E. | | Deposit date: | 2020-10-20 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex.
Elife, 9, 2020
|
|
7AJR
 
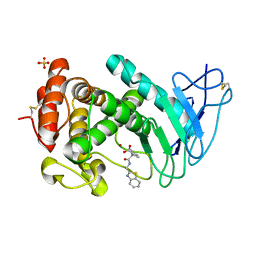 | | Virtual screening approach leading to the identification of a novel and tractable series of Pseudomonas aeruginosa elastase inhibitors | | Descriptor: | 2-[2-(1,3-benzothiazol-2-ylmethylcarbamoyl)-1,3-dihydroinden-2-yl]ethanoic acid, Keratinase KP2, SULFATE ION, ... | | Authors: | Leiris, S, Davies, D.T, Sprinsky, N, Castandet, J, Behria, L, Bodnarchuk, M.S, Sutton, J.M, Mullins, T.M.G, Jones, M.W, Forrest, A.K, Pallin, T.D, Karunakar, P, Martha, S.K, Parusharamulu, B, Ramula, R, Kotha, V, Pottabathini, N, Pothukanuri, S, Lemonnier, M, Everett, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual Screening Approach to Identifying a Novel and Tractable Series of Pseudomonas aeruginosa Elastase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
6IF7
 
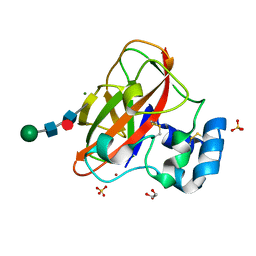 | | Crystal Structure of AA10 Lytic Polysaccharide Monooxygenase from Tectaria macrodonta | | Descriptor: | COPPER (II) ION, Chitin binding protein, GLYCEROL, ... | | Authors: | Archana, A, Yadav, S.K, Singh, P.K, Vasudev, P.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insecticidal fern protein Tma12 is possibly a lytic polysaccharide monooxygenase.
Planta, 249, 2019
|
|
6YSV
 
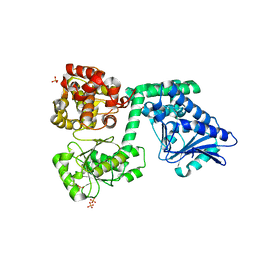 | |
6YSW
 
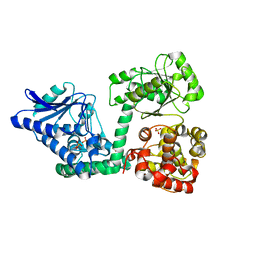 | | E. coli anaerobic trifunctional enzyme subunit-alpha in complex with coenzyme A | | Descriptor: | COENZYME A, Fatty acid oxidation complex subunit alpha, SULFATE ION | | Authors: | Sah-Teli, S.K, Hynonen, M.J, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2020-04-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes
Structure, 2023
|
|
5FG0
 
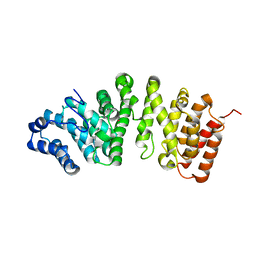 | |
5FT2
 
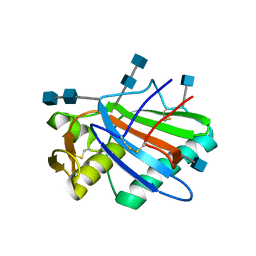 | | Sub-tomogram averaging of Lassa virus glycoprotein spike from virus- like particles at pH 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRE-GLYCOPROTEIN POLYPROTEIN GP COMPLEX | | Authors: | Li, S, Zhaoyang, S, Pryce, R, Parsy, M.L, Fehling, S.K, Schlie, K, Siebert, C.A, Garten, W, Bowden, T.A, Strecker, T, Huiskonen, J.T. | | Deposit date: | 2016-01-09 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Acidic Ph-Induced Conformations and Lamp1 Binding of the Lassa Virus Glycoprotein Spike.
Plos Pathog., 12, 2016
|
|
7RTY
 
 | |
3RO6
 
 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg ion | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative chloromuconate cycloisomerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-25 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3R03
 
 | |
3R9E
 
 | |
7SJ4
 
 | | Human Trio residues 1284-1959 in complex with Rac1 | | Descriptor: | Ras-related C3 botulinum toxin substrate 1, Triple functional domain protein | | Authors: | Chen, C.-L, Ravala, S.K, Bandekar, S.J, Cash, J, Tesmer, J.J.G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structural/functional studies of Trio provide insights into its configuration and show that conserved linker elements enhance its activity for Rac1.
J.Biol.Chem., 298, 2022
|
|
8BRJ
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) trimeric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
8BNU
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) tetrameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
8BNR
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) octameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (10.3 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
