6A9U
 
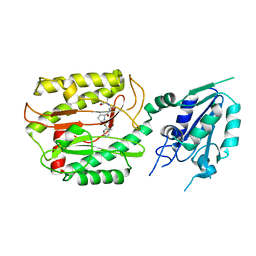 | | Crystal strcture of Icp55 from Saccharomyces cerevisiae bound to apstatin inhibitor | | Descriptor: | Intermediate cleaving peptidase 55, MANGANESE (II) ION, apstatin | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9V
 
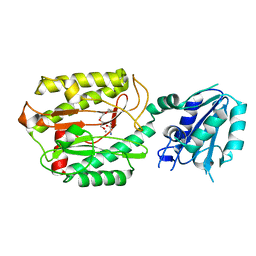 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9T
 
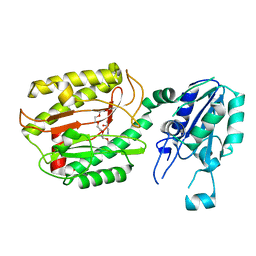 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 58 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
5CFY
 
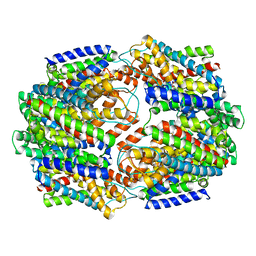 | | CRYSTAL STRUCTURE OF GLTPH R397A IN COMPLEX WITH NA+ AND L-ASP | | Descriptor: | 425aa long hypothetical proton glutamate symport protein, ASPARTIC ACID, SODIUM ION | | Authors: | Boudker, O, Oh, S. | | Deposit date: | 2015-07-08 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Coupled ion binding and structural transitions along the transport cycle of glutamate transporters.
Elife, 3, 2014
|
|
7DFS
 
 | | Crystal structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S - Rha-GlcA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase, alpha-D-mannopyranose, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7DFQ
 
 | | Crystal Structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S, ligand-free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7DBN
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
7DBM
 
 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
6L0L
 
 | | Hydra-1ubq de nova designed by Hydra based on ubiquitin | | Descriptor: | Hydra-1ubq | | Authors: | Ouyang, B. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiobjective heuristic algorithm for de novo protein design in a quantified continuous sequence space.
Comput Struct Biotechnol J, 19, 2021
|
|
6LCE
 
 | |
6LCF
 
 | | Crystal Structure of beta-L-arabinobiose binding protein - native | | Descriptor: | ABC transporter substrate binding component, beta-L-arabinofuranose-(1-2)-beta-L-arabinofuranose | | Authors: | Miyake, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of beta-L-arabinobiose-binding protein in the metabolic pathway of hydroxyproline-rich glycoproteins in Bifidobacterium longum.
Febs J., 287, 2020
|
|
7F3Y
 
 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with methotrexate (MTX), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | Authors: | Vanichtanankul, J, Tanramluk, D, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
7F3Z
 
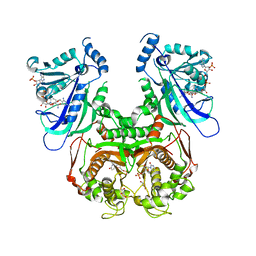 | | Double mutant Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS-K1, C59R+S108N) complexed with Trimethoprim (TOP), NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, GLYCEROL, ... | | Authors: | Vanichtanankul, J, Tanramluk, D, Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | MANORAA: A machine learning platform to guide protein-ligand design by anchors and influential distances.
Structure, 30, 2022
|
|
6MKM
 
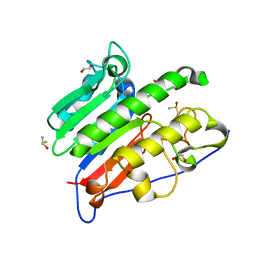 | | Crystallographic solvent mapping analysis of DMSO/Tris bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MK3
 
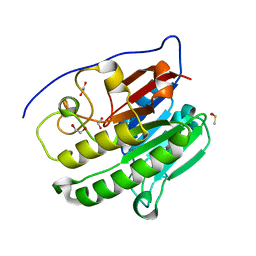 | | Crystallographic solvent mapping analysis of DMSO bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MKO
 
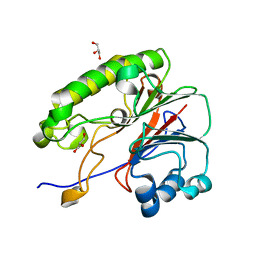 | |
6MKK
 
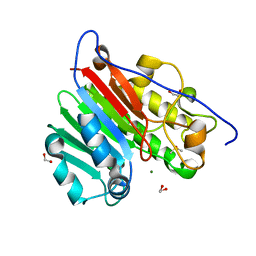 | | Crystallographic solvent mapping analysis of DMSO/Mg bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
7JH4
 
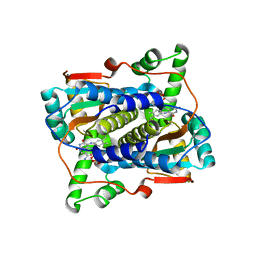 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
4ALZ
 
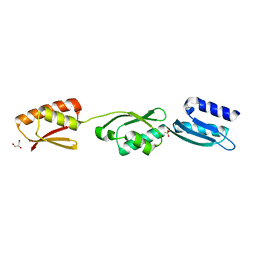 | | The Yersinia T3SS basal body component YscD reveals a different structural periplasmatic domain organization to known homologue PrgH | | Descriptor: | GLYCEROL, PHOSPHATE ION, YOP PROTEINS TRANSLOCATION PROTEIN D | | Authors: | Schmelz, S, Wisand, U, Stenta, M, Muenich, S, Widow, U, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2012-03-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | In Situ Structural Analysis of the Yersinia Enterocolitica Injectisome.
Elife, 2, 2013
|
|
4B08
 
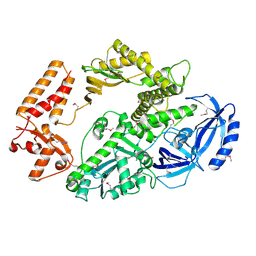 | | Yeast DNA polymerase alpha, Selenomethionine protein | | Descriptor: | DNA POLYMERASE ALPHA CATALYTIC SUBUNIT A | | Authors: | Perera, R.L, Torella, R, Klinge, S, Kilkenny, M.L, Maman, J.D, Pellegrini, L. | | Deposit date: | 2012-06-29 | | Release date: | 2013-02-27 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Mechanism for Priming DNA Synthesis by Yeast DNA Polymerase Alpha
Elife, 2, 2013
|
|
4B8V
 
 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
4B9H
 
 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer: I3C heavy atom derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
4BL0
 
 | | Crystal structure of yeast Bub3-Bub1 bound to phospho-Spc105 | | Descriptor: | CELL CYCLE ARREST PROTEIN BUB3, CHECKPOINT SERINE/THREONINE-PROTEIN KINASE BUB1, MAGNESIUM ION, ... | | Authors: | Primorac, I, Weir, J.R, Musacchio, A. | | Deposit date: | 2013-04-30 | | Release date: | 2013-09-18 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bub3 Reads Phosphorylated Melt Repeats to Promote Spindle Assembly Checkpoint Signaling
Elife, 2, 2013
|
|
4BZI
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, SAR1P, ... | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
4BZK
 
 | | The structure of the COPII coat assembled on membranes | | Descriptor: | Protein transport protein SEC13, Protein transport protein SEC31 | | Authors: | Zanetti, G, Prinz, S, Daum, S, Meister, A, Schekman, R, Bacia, K, Briggs, J.A.G. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (40 Å) | | Cite: | The Structure of the Copii Transport-Vesicle Coat Assembled on Membranes
Elife, 2, 2013
|
|
