2EDT
 
 | | Solution structure of the ig-like domain (3449-3537) from human Obscurin | | Descriptor: | Obscurin | | Authors: | Seimiya, K, Izumi, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ig-like domain (3449-3537) from human Obscurin
to be published
|
|
2EO1
 
 | | Solution structure of the ig domain of human OBSCN protein | | Descriptor: | CDNA FLJ14124 fis, clone MAMMA1002498 | | Authors: | Nagashima, K, Nagashima, T, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ig domain of human OBSCN protein
To be Published
|
|
2AVG
 
 | |
2EDQ
 
 | |
2E7B
 
 | |
2EDJ
 
 | | Solution structure of the fifth ig-like domain from human Roundabout homolog 2 | | Descriptor: | Roundabout homolog 2 | | Authors: | Kurosaki, C, Izumi, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2007-08-14 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fifth ig-like domain from human Roundabout homolog 2
to be published
|
|
2EDR
 
 | | Solution structure of the ig-like domain (3361-3449) of human obscurin | | Descriptor: | Obscurin | | Authors: | Kurosaki, C, Izumi, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ig-like domain (3361-3449) of human obscurin
To be Published
|
|
2EDF
 
 | |
2EDW
 
 | |
2ENY
 
 | | Solution structure of the ig-like domain (2735-2825) of human obscurin | | Descriptor: | Obscurin | | Authors: | Nagashima, K, Izumi, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ig-like domain (2735-2825) of human obscurin
To be Published
|
|
2DM2
 
 | |
2CR6
 
 | |
4XB8
 
 | | Crystal structure of Dscam1 isoform 9.44, N-terminal four Ig domains (with zinc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Down Syndrome Cell Adhesion Molecule, ... | | Authors: | Chen, Q, Yu, Y, Li, S.A, cheng, L. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural basis of Dscam1 homodimerization: Insights into context constraint for protein recognition
Sci Adv, 2, 2016
|
|
6F2O
 
 | |
1QZ1
 
 | | Crystal Structure of the Ig 1-2-3 fragment of NCAM | | Descriptor: | Neural cell adhesion molecule 1, 140 kDa isoform | | Authors: | Soroka, V, Kolkova, K, Kastrup, J.S, Diederichs, K, Breed, J, Kiselyov, V.V, Poulsen, F.M, Larsen, I.K, Welte, W, Berezin, V, Bock, E, Kasper, C. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion
Structure, 11, 2003
|
|
1BIH
 
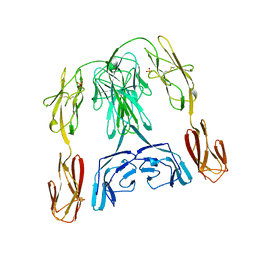 | | CRYSTAL STRUCTURE OF THE INSECT IMMUNE PROTEIN HEMOLIN: A NEW DOMAIN ARRANGEMENT WITH IMPLICATIONS FOR HOMOPHILIC ADHESION | | Descriptor: | HEMOLIN, PHOSPHATE ION | | Authors: | Su, X.-D, Gastinel, L.N, Vaughn, D.E, Faye, I, Poon, P, Bjorkman, P.J. | | Deposit date: | 1998-06-17 | | Release date: | 1998-10-14 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of hemolin: a horseshoe shape with implications for homophilic adhesion.
Science, 281, 1998
|
|
6U6T
 
 | | Neuronal growth regulator 1 (NEGR1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuronal growth regulator 1, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G. | | Deposit date: | 2019-08-30 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
4KC3
 
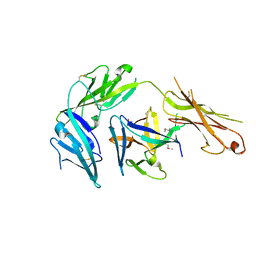 | | Cytokine/receptor binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor-like 1, Interleukin-33 | | Authors: | Liu, X, Wang, X.Q. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2702 Å) | | Cite: | Structural insights into the interaction of IL-33 with its receptors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
9BA5
 
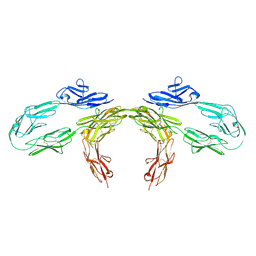 | |
6S9F
 
 | | Drosophila OTK, extracellular domains 3-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tyrosine-protein kinase-like otk | | Authors: | Rozbesky, D, Jones, E.Y. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Drosophila OTK Is a Glycosaminoglycan-Binding Protein with High Conformational Flexibility.
Structure, 28, 2020
|
|
3JXA
 
 | | Immunoglobulin domains 1-4 of mouse CNTN4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin 4 | | Authors: | Bouyain, S. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5XNP
 
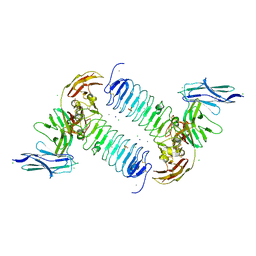 | | Crystal structures of human SALM5 in complex with human PTPdelta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, H, Lin, Z, Xu, F. | | Deposit date: | 2017-05-24 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.729 Å) | | Cite: | Structural basis of SALM5-induced PTP delta dimerization for synaptic differentiation
Nat Commun, 9, 2018
|
|
5XWU
 
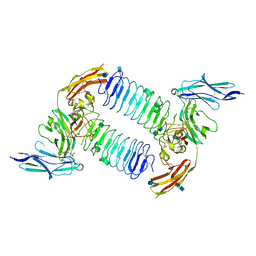 | | Crystal structure of PTPdelta Ig1-Ig3 in complex with SALM2 LRR-Ig | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
5E4I
 
 | | Crystal structure of mouse CNTN5 Ig1-Ig4 domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-5 | | Authors: | Nikolaienko, R.M, Bouyain, S. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Interactions Between Contactin Family Members and Protein-tyrosine Phosphatase Receptor Type G in Neural Tissues.
J.Biol.Chem., 291, 2016
|
|
3LAF
 
 | | Structure of DCC, a netrin-1 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Deleted in Colorectal Cancer, SULFATE ION, ... | | Authors: | Chen, Q, Liu, J.-H, Wang, J.-H. | | Deposit date: | 2010-01-06 | | Release date: | 2011-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | N-terminal horseshoe conformation of DCC is functionally required for axon guidance and might be shared by other neural receptors.
J.Cell.Sci., 126, 2013
|
|
