4MLP
 
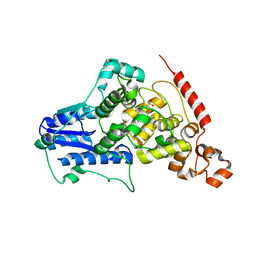 | |
4K0R
 
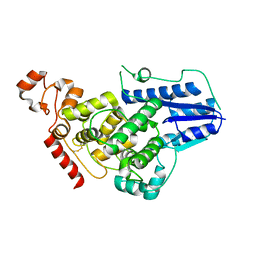 | | Crystal structure of mouse Cryptochrome 1 | | Descriptor: | Cryptochrome-1 | | Authors: | Czarna, A, Wolf, E. | | Deposit date: | 2013-04-04 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
4JZY
 
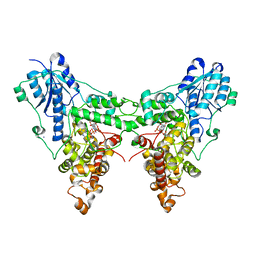 | | Crystal structures of Drosophila Cryptochrome | | Descriptor: | AMMONIUM ION, Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Czarna, A, Wolf, E. | | Deposit date: | 2013-04-03 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K03
 
 | | Crystal structure of Drosophila Cryprochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Berndt, A, Wolf, E. | | Deposit date: | 2013-04-03 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I6G
 
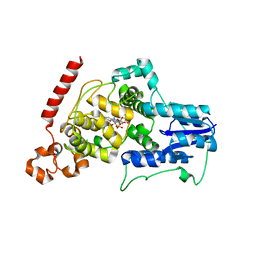 | | a vertebrate cryptochrome with FAD | | Descriptor: | Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
4I6E
 
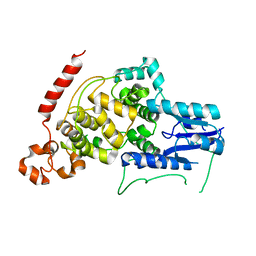 | | A vertebrate cryptochrome | | Descriptor: | Cryptochrome-2 | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
4I6J
 
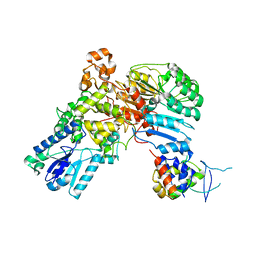 | | A ubiquitin ligase-substrate complex | | Descriptor: | Cryptochrome-2, F-box/LRR-repeat protein 3, S-phase kinase-associated protein 1 | | Authors: | Xing, W, Busino, L, Hinds, T.R, Marionni, S.T, Saifee, N.H, Bush, M.F, Pagano, M, Zheng, N. | | Deposit date: | 2012-11-29 | | Release date: | 2013-03-13 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SCFFBXL3 ubiquitin ligase targets cryptochromes at their cofactor pocket.
Nature, 496, 2013
|
|
4GU5
 
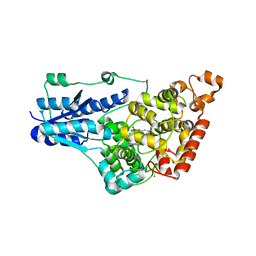 | | Structure of Full-length Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zoltowski, B.D, Vaidya, A.T, Top, D, Widom, J, Young, M.W, Levy, C, Jones, A.R, Scrutton, N.S, Leys, D, Crane, B.R. | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Updated structure of Drosophila cryptochrome.
Nature, 495, 2013
|
|
2WQ7
 
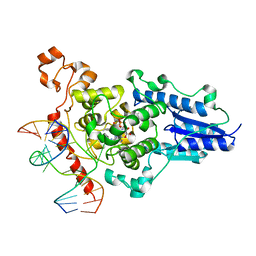 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(6-4)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*ZP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions.
J.Am.Chem.Soc., 132, 2010
|
|
2WQ6
 
 | | Structure of the 6-4 photolyase of D. melanogaster in complex with the non-natural N4-methyl T(Dewar)C lesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*TDYP*CDWP*GP* CP*AP*AP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP* CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Kaya, E, Schneider, S, Maul, M.J, Carell, T. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions
J.Am.Chem.Soc., 132, 2010
|
|
2WB2
 
 | | Drosophila Melanogaster (6-4) Photolyase Bound To double stranded Dna containing a T(6-4)C Photolesion | | Descriptor: | 5'-D(*AP*CP*AP*GP*CP*GP*GP*64PP*ZP*GP*CP*AP *GP*GP*T)-3', 5'-D(*TP*AP*CP*CP*TP*GP*CP*GP*AP*CP*CP*GP*CP*TP*G)-3', FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Glas, A.F, Schneider, S, Maul, M.J, Hennecke, U, Carell, T. | | Deposit date: | 2009-02-20 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the T(6-4)C Lesion in Complex with a (6-4) DNA Photolyase and Repair of Uv- Induced (6-4) and Dewar Photolesions.
Chemistry, 15, 2009
|
|
3FY4
 
 | | (6-4) Photolyase Crystal Structure | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-4 photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2009-01-21 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2VTB
 
 | | Structure of cryptochrome 3 - DNA complex | | Descriptor: | 5'-D(*DT*DT*DT*DT*DTP)-3', 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, ACETATE ION, ... | | Authors: | Pokorny, R, Klar, T, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2008-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Recognition and Repair of Uv Lesions in Loop Structures of Duplex DNA by Dash-Type Cryptochrome.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CVU
 
 | | Drosophila melanogaster (6-4) photolyase bound to ds DNA with a T-T (6-4) photolesion | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and mechanism of a DNA (6-4) photolyase.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
3CVW
 
 | | Drosophila melanogaster (6-4) photolyase H365N mutant bound to ds DNA with a T-T (6-4) photolesion and cofactor F0 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a cofactor f0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
3CVX
 
 | | Drosophila melanogaster (6-4) photolyase H369M mutant bound to ds DNA with a T-T (6-4) photolesion | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a coenzyme F0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
3CVV
 
 | | Drosophila melanogaster (6-4) photolyase bound to ds DNA with a T-T (6-4) photolesion and F0 cofactor | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), ... | | Authors: | Glas, A.F, Maul, M.J, Cryle, M.J, Barends, T.R.M, Schneider, S, Kaya, E, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The archaeal cofactor F0 is a light-harvesting antenna chromophore in eukaryotes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3CVY
 
 | | Drosophila melanogaster (6-4) photolyase bound to repaired ds DNA | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*DTP*DTP*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and mechanism of a DNA (6-4) photolyase.
Angew.Chem.Int.Ed.Engl., 47, 2008
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
2IJG
 
 | | Crystal Structure of cryptochrome 3 from Arabidopsis thaliana | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, Cryptochrome DASH, chloroplast/mitochondrial, ... | | Authors: | Huang, Y, Deisenhofer, J. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2J4D
 
 | | Cryptochrome 3 from Arabidopsis thaliana | | Descriptor: | 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, CRYPTOCHROME DASH, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Klar, T, Pokorny, R, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-28 | | Release date: | 2007-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cryptochrome 3 from Arabidopsis Thaliana: Structural and Functional Analysis of its Complex with a Folate Light Antenna
J.Mol.Biol., 366, 2007
|
|
2J07
 
 | | Thermus DNA photolyase with 8-HDF antenna chromophore | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J09
 
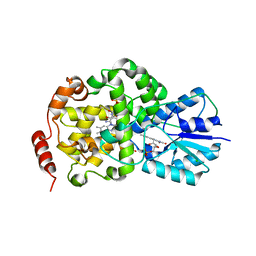 | | Thermus DNA photolyase with FMN antenna chromophore | | Descriptor: | CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
2J08
 
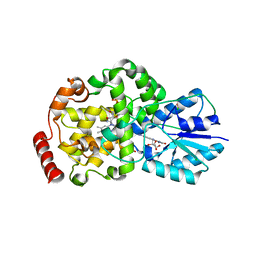 | | Thermus DNA photolyase with 8-Iod-riboflavin antenna chromophore | | Descriptor: | 1-DEOXY-1-(8-IODO-7-METHYL-2,4-DIOXO-3,4-DIHYDROBENZO[G]PTERIDIN-10(2H)-YL)-D-RIBITOL, CHLORIDE ION, DEOXYRIBODIPYRIMIDINE PHOTO-LYASE, ... | | Authors: | Klar, T, Kaiser, G, Hennecke, U, Carell, T, Batschauer, A, Essen, L.-O. | | Deposit date: | 2006-08-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Natural and Non-Natural Antenna Chromophores in the DNA Photolyase from Thermus Thermophilus
Chembiochem, 7, 2006
|
|
1U3C
 
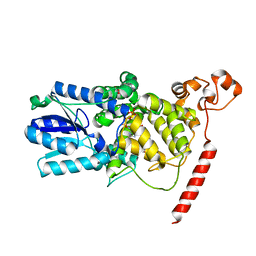 | | Crystal Structure of the PHR domain of Cryptochrome 1 from Arabidopsis thaliana | | Descriptor: | CHLORIDE ION, Cryptochrome 1 apoprotein, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, ... | | Authors: | Brautigam, C.A, Smith, B.S, Ma, Z, Palnitkar, M, Tomchick, D.R, Machius, M, Deisenhofer, J. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
