2E1H
 
 | |
2E10
 
 | |
2DZC
 
 | | Crystal Structure Of Biotin Protein Ligase From Pyrococcus Horikoshii, Mutation R48A | | Descriptor: | biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | Bagautdinov, B, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-27 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
2DZ9
 
 | |
2DXU
 
 | | Crystal Structure Of Biotin Protein Ligase From Pyrococcus Horikoshii Complexed with Biotinyl-5'-AMP, Mutation R48A | | Descriptor: | BIOTINYL-5-AMP, biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | Bagautdinov, B, Taketa, M, Matsuura, Y, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-30 | | Release date: | 2007-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
2DXT
 
 | |
2DVE
 
 | |
2HNI
 
 | |
2DTO
 
 | |
2DTI
 
 | |
2DTH
 
 | |
2DKG
 
 | |
2DJZ
 
 | |
2CGH
 
 | |
2DEQ
 
 | |
2FYK
 
 | |
2C8M
 
 | |
2C7I
 
 | |
2EWN
 
 | | Ecoli Biotin Repressor with co-repressor analog | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, BirA bifunctional protein | | Authors: | Wood, Z.A, Weaver, L.H, Matthews, B.W. | | Deposit date: | 2005-11-04 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Co-repressor Induced Order and Biotin Repressor Dimerization: A Case for Divergent Followed by Convergent Evolution.
J.Mol.Biol., 357, 2006
|
|
2ARU
 
 | | Crystal structure of lipoate-protein ligase A bound with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
2ARS
 
 | | Crystal structure of lipoate-protein ligase A From Thermoplasma acidophilum | | Descriptor: | Lipoate-protein ligase A, MAGNESIUM ION | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
2ART
 
 | | Crystal structure of lipoate-protein ligase A bound with lipoyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, LIPOIC ACID, Lipoate-protein ligase A, ... | | Authors: | Kim, D.J, Kim, K.H, Lee, H.H, Lee, S.J, Ha, J.Y, Yoon, H.J, Suh, S.W. | | Deposit date: | 2005-08-22 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains
J.Biol.Chem., 280, 2005
|
|
1X2H
 
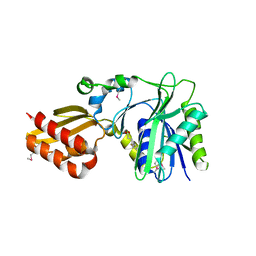 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli complexed with lipoic acid | | Descriptor: | LIPOIC ACID, Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1X2G
 
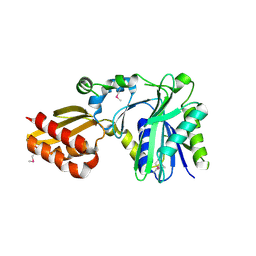 | | Crystal Structure of Lipate-Protein Ligase A from Escherichia coli | | Descriptor: | Lipoate-protein ligase A | | Authors: | Fujiwara, K, Toma, S, Okamura-Ikeda, K, Motokawa, Y, Nakagawa, A, Taniguchi, H. | | Deposit date: | 2005-04-23 | | Release date: | 2005-08-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site
J.Biol.Chem., 280, 2005
|
|
1X01
 
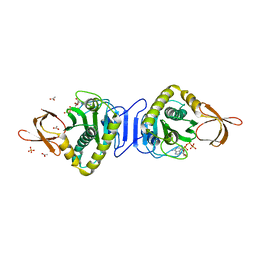 | |
