6AMS
 
 | |
6AMQ
 
 | |
8VAS
 
 | |
8VAQ
 
 | |
8VAT
 
 | |
8VAL
 
 | |
8VAM
 
 | |
8VAR
 
 | |
8VAP
 
 | |
5FKU
 
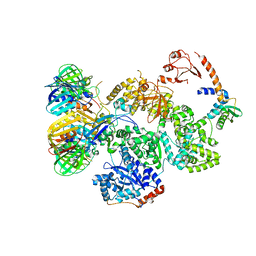 | | cryo-EM structure of the E. coli replicative DNA polymerase complex in DNA free state (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III SUBUNIT ALPHA, DNA POLYMERASE III SUBUNIT BETA, DNA POLYMERASE III SUBUNIT EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.34 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FRQ
 
 | |
5FKW
 
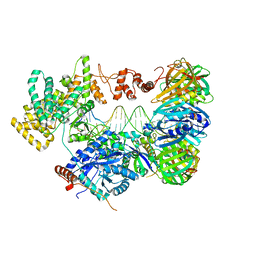 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon) | | Descriptor: | DNA POLYMERASE III ALPHA, DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FXT
 
 | |
3P16
 
 | | Crystal structure of DNA polymerase III sliding clamp | | Descriptor: | DNA polymerase III subunit beta | | Authors: | Gui, W.J, Lin, S.Q, Chen, Y.Y, Zhang, X.E, Bi, L.J, Jiang, T. | | Deposit date: | 2010-09-30 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of DNA polymerase III beta sliding clamp from Mycobacterium tuberculosis.
Biochem.Biophys.Res.Commun., 405, 2011
|
|
5FKV
 
 | | cryo-EM structure of the E. coli replicative DNA polymerase complex bound to DNA (DNA polymerase III alpha, beta, epsilon, tau complex) | | Descriptor: | DNA POLYMERASE III BETA, DNA POLYMERASE III EPSILON, DNA POLYMERASE III SUBUNIT ALPHA, ... | | Authors: | Fernandez-Leiro, R, Conrad, J, Scheres, S.H.W, Lamers, M.H. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.04 Å) | | Cite: | cryo-EM structures of theE. colireplicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease andtau.
Elife, 4, 2015
|
|
5FVE
 
 | |
3Q4J
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
5G4Q
 
 | | H.pylori Beta clamp in complex with 5-chloroisatin | | Descriptor: | 5-chloro-1H-indole-2,3-dione, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-16 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening of E. coli beta-clamp Inhibitors Revealed that Few Inhibit Helicobacter pylori More Effectively: Structural and Functional Characterization.
Antibiotics (Basel), 7, 2018
|
|
5G48
 
 | | H.pylori Beta clamp in complex with Diflunisal | | Descriptor: | 5-(2,4-DIFLUOROPHENYL)-2-HYDROXY-BENZOIC ACID, DNA POLYMERASE III SUBUNIT BETA | | Authors: | Pandey, P, Gourinath, S. | | Deposit date: | 2016-05-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Targeting the beta-clamp in Helicobacter pylori with FDA-approved drugs reveals micromolar inhibition by diflunisal.
FEBS Lett., 591, 2017
|
|
3Q4K
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3Q4L
 
 | | Structure of a small peptide ligand bound to E.coli DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION, peptide ligand | | Authors: | Wolff, P, Olieric, V, Briand, J.P, Chaloin, O, Dejaegere, A, Dumas, P, Ennifar, E, Guichard, G, Wagner, J, Burnouf, D. | | Deposit date: | 2010-12-23 | | Release date: | 2011-12-28 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of short peptide ligands binding onto the E. coli processivity ring.
J.Med.Chem., 54, 2011
|
|
3PWE
 
 | | Crystal structure of the E. coli beta clamp mutant R103C, I305C, C260S, C333S at 2.2A resolution | | Descriptor: | DNA polymerase III subunit beta | | Authors: | Marzahn, M.R, Robbins, A.H, McKenna, R, Bloom, L.B. | | Deposit date: | 2010-12-08 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The E. coli clamp loader can actively pry open the beta-sliding clamp
J.Biol.Chem., 286, 2011
|
|
3QSB
 
 | | Structure of E. coli polIIIbeta with (Z)-5-(1-((4'-Fluorobiphenyl-4-yl)methoxyimino)butyl)-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile | | Descriptor: | (1R,5R)-5-{(1Z)-N-[(4'-fluorobiphenyl-4-yl)methoxy]butanimidoyl}-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile, DNA polymerase III subunit beta | | Authors: | Wijffels, G, Johnson, W.M, Oakley, A.J, Turner, K, Epa, V.C, Briscoe, S.J, Polley, M, Liepa, A.J, Hofmann, A, Buchardt, J, Christensen, C, Prosselkov, P, Dalrymple, B.P, Alewood, P.F, Jennings, P.A, Dixon, N.E, Winkler, D.A. | | Deposit date: | 2011-02-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding inhibitors of the bacterial sliding clamp by design
J.Med.Chem., 54, 2011
|
|
3RB9
 
 | |
3T0P
 
 | |
