3EZX
 
 | |
3IV9
 
 | |
3IVA
 
 | | Structure of the B12-dependent Methionine Synthase (MetH) C-teminal half with AdoHcy bound | | Descriptor: | COBALAMIN, Methionine synthase, NITRATE ION, ... | | Authors: | Pattridge, K.A, Koutmos, M, Smith, J.L. | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the reactivation of cobalamin-dependent methionine synthase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KP1
 
 | | Crystal structure of ornithine 4,5 aminomutase (Resting State) | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOW
 
 | | Crystal Structure of ornithine 4,5 aminomutase backsoaked complex | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOX
 
 | | Crystal Structure of ornithine 4,5 aminomutase in complex with 2,4-diaminobutyrate (Anaerobic) | | Descriptor: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOZ
 
 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Anaerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KP0
 
 | | Crystal Structure of ORNITHINE 4,5 AMINOMUTASE in complex with 2,4-diaminobutyrate (DAB) (Aerobic) | | Descriptor: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOY
 
 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Aerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
2XIQ
 
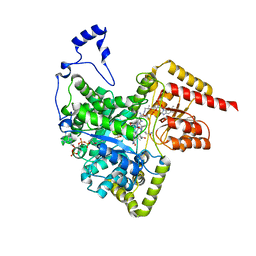 | | Crystal structure of human methylmalonyl-CoA mutase in complex with adenosylcobalamin and malonyl-CoA | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, MALONYL-COENZYME A, ... | | Authors: | Yue, W.W, Froese, D.S, Kochan, G, Chaikuad, A, Krojer, T, Muniz, J, Ugochukwu, E, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Human Gtpase Mmaa and Vitamin B12-Dependent Methylmalonyl-Coa Mutase and Insight Into Their Complex Formation.
J.Biol.Chem., 285, 2010
|
|
2XIJ
 
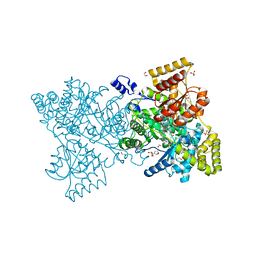 | | Crystal structure of human methylmalonyl-CoA mutase in complex with adenosylcobalamin | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-DEOXYADENOSINE, ... | | Authors: | Yue, W.W, Froese, D.S, Kochan, G, Chaikuad, A, Krojer, T, Muniz, J, Vollmar, M, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Human Gtpase Mmaa and Vitamin B12-Dependent Methylmalonyl-Coa Mutase and Insight Into Their Complex Formation.
J.Biol.Chem., 285, 2010
|
|
4JGI
 
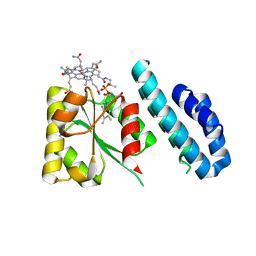 | | 1.5 Angstrom crystal structure of a novel cobalamin-binding protein from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CO-METHYLCOBALAMIN, Putative uncharacterized protein | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-03-01 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the cobalamin-binding protein of a putative O-demethylase from Desulfitobacterium hafniense DCB-2.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4Q37
 
 | |
4R3U
 
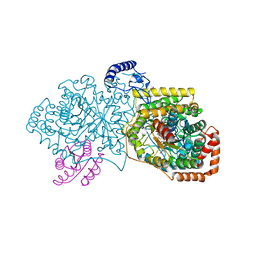 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Mutase | | Descriptor: | 2-hydroxyisobutyryl-CoA mutase large subunit, 2-hydroxyisobutyryl-CoA mutase small subunit, 3-HYDROXYBUTANOYL-COENZYME A, ... | | Authors: | Zahn, M, Kurteva-Yaneva, N, Rohwerder, T, Straeter, N. | | Deposit date: | 2014-08-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the stereospecificity of bacterial B12-dependent 2-hydroxyisobutyryl-CoA mutase.
J.Biol.Chem., 290, 2015
|
|
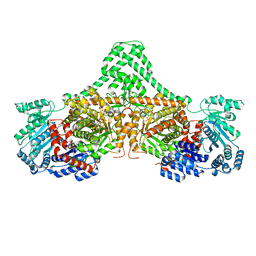
4XC7
 
 | |
4XC6
 
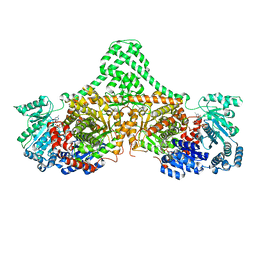 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, and Mg (holo-IcmF/GDP) | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XC8
 
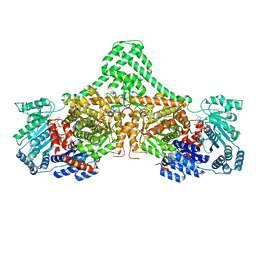 | | Isobutyryl-CoA mutase fused with bound butyryl-CoA, GDP, and Mg and without cobalamin (apo-IcmF/GDP) | | Descriptor: | Butyryl Coenzyme A, GUANOSINE-5'-DIPHOSPHATE, Isobutyryl-CoA mutase fused, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2014-12-17 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Visualization of a radical B12 enzyme with its G-protein chaperone.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CJV
 
 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, Mg (holo-IcmF/GDP), and substrate isovaleryl-coenzyme A | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Basis for Substrate Specificity in Adenosylcobalamin-dependent Isobutyryl-CoA Mutase and Related Acyl-CoA Mutases.
J.Biol.Chem., 290, 2015
|
|
5CJT
 
 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, Mg (holo-IcmF/GDP), and substrate isobutyryl-coenzyme A | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for Substrate Specificity in Adenosylcobalamin-dependent Isobutyryl-CoA Mutase and Related Acyl-CoA Mutases.
J.Biol.Chem., 290, 2015
|
|
5CJU
 
 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, Mg (holo-IcmF/GDP), and substrate n-butyryl-coenzyme A | | Descriptor: | 5'-DEOXYADENOSINE, Butyryl Coenzyme A, COBALAMIN, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis for Substrate Specificity in Adenosylcobalamin-dependent Isobutyryl-CoA Mutase and Related Acyl-CoA Mutases.
J.Biol.Chem., 290, 2015
|
|
5CJW
 
 | | Isobutyryl-CoA mutase fused with bound adenosylcobalamin, GDP, Mg (holo-IcmF/GDP), and substrate pivalyl-coenzyme A | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jost, M, Drennan, C.L. | | Deposit date: | 2015-07-15 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for Substrate Specificity in Adenosylcobalamin-dependent Isobutyryl-CoA Mutase and Related Acyl-CoA Mutases.
J.Biol.Chem., 290, 2015
|
|
6H9F
 
 | | Structure of glutamate mutase reconstituted with bishomo-coenzyme B12 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-propyl-oxolane-3,4-diol, COBALAMIN, D(-)-TARTARIC ACID, ... | | Authors: | Gruber, K, Csitkovits, V, Kratky, C. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Demystification of Radical Catalysis by a Coenzyme B 12 Dependent Enzyme-Crystallographic Study of Glutamate Mutase with Cofactor Homologues.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6H9E
 
 | | Structure of glutamate mutase reconstituted with homo-coenzyme B12 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, COBALAMIN, D(-)-TARTARIC ACID, ... | | Authors: | Gruber, K, Csitkovits, V, Kratky, C. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Based Demystification of Radical Catalysis by a Coenzyme B 12 Dependent Enzyme-Crystallographic Study of Glutamate Mutase with Cofactor Homologues.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6OXD
 
 | | Structure of Mycobacterium tuberculosis methylmalonyl-CoA mutase with adenosyl cobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Itaconyl coenzyme A, ... | | Authors: | Purchal, M, Ruetz, M, Banerjee, R, Koutmos, M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair.
Science, 366, 2019
|
|
6OXC
 
 | | Structure of Mycobacterium tuberculosis methylmalonyl-CoA mutase with adenosyl cobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Methylmalonyl-CoA mutase large subunit, ... | | Authors: | Purchal, M, Ruetz, M, Banerjee, R, Koutmos, M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair.
Science, 366, 2019
|
|
