7CF6
 
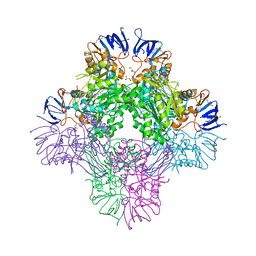 | | Crystal structure of Beta-aspartyl dipeptidase from thermophilic keratin degrading Fervidobacterium islandicum AW-1 in complex with beta-Asp-Leu dipeptide | | Descriptor: | (2S)-2-[[(3S)-3-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]amino]-4-methyl-pentanoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dhanasingh, I, La, J.W, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium islandicum AW-1.
Front Mol Biosci, 7, 2020
|
|
7LKK
 
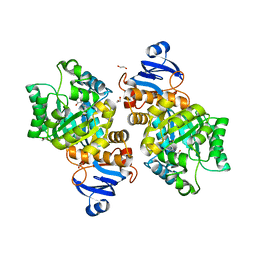 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) in complex with Methylthio-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, 1,2-ETHANEDIOL, Aminofutalosine deaminase, ... | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
7LKJ
 
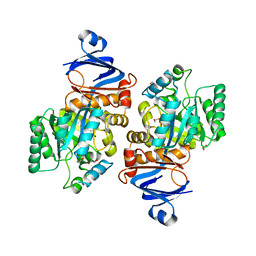 | | Crystal structure of Helicobacter pylori aminofutalosine deaminase (AFLDA) | | Descriptor: | 1,2-ETHANEDIOL, Aminofutalosine deaminase, FE (III) ION | | Authors: | Harijan, R.K, Feng, M, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Aminofutalosine Deaminase in the Menaquinone Pathway of Helicobacter pylori .
Biochemistry, 60, 2021
|
|
3GRI
 
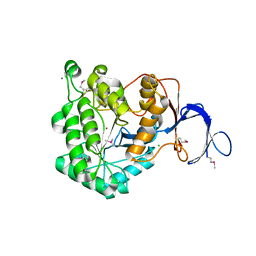 | | The Crystal Structure of a Dihydroorotase from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-25 | | Release date: | 2009-05-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of a Dihydroorotase from Staphylococcus aureus
To be Published
|
|
3HM7
 
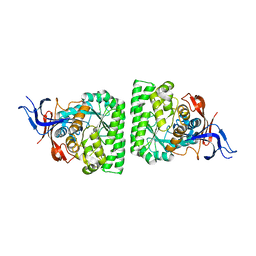 | | Crystal structure of allantoinase from Bacillus halodurans C-125 | | Descriptor: | Allantoinase, ZINC ION | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Miller, S, Wasserman, S.R, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Allantoinase from Bacillus Halodurans
To be Published
|
|
6OHC
 
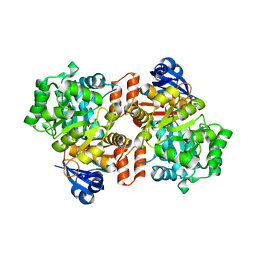 | | E. coli Guanine Deaminase | | Descriptor: | GLYCEROL, Guanine deaminase, ZINC ION | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
6OH9
 
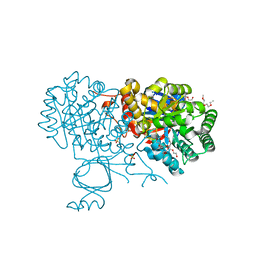 | | Yeast Guanine Deaminase | | Descriptor: | NONAETHYLENE GLYCOL, SULFATE ION, Yeast Guanine Deaminase, ... | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
6OHB
 
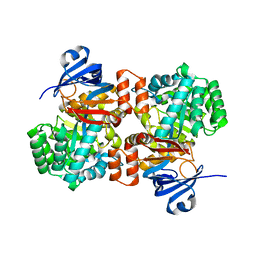 | | E. coli Guanine Deaminase | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
3IV8
 
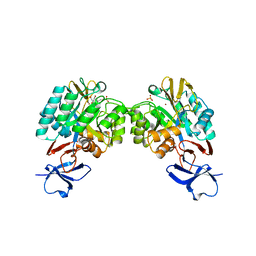 | | N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae complexed with fructose 6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, N-acetylglucosamine-6-phosphate deacetylase, NICKEL (II) ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | X-ray crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae complexed with fructose 6-phosphate.
To be Published
|
|
6OHA
 
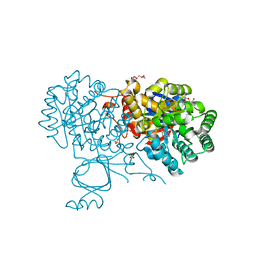 | | Yeast Guanine Deaminase | | Descriptor: | PENTAETHYLENE GLYCOL, Probable guanine deaminase, SULFATE ION, ... | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
3JZE
 
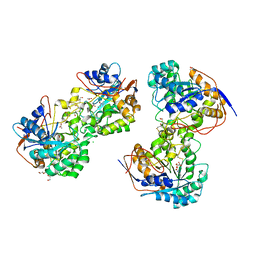 | | 1.8 Angstrom resolution crystal structure of dihydroorotase (pyrC) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ACETIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Dihydroorotase (pyrC) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2.
TO BE PUBLISHED
|
|
7U5K
 
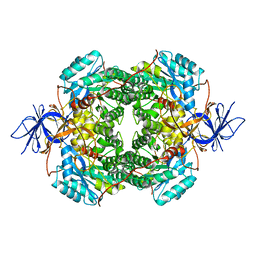 | | Cryo-EM Structure of DPYSL2 | | Descriptor: | Dihydropyrimidinase-related protein 2 | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
4RDW
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutaric acid
To be Published
|
|
7UOF
 
 | | Dihydroorotase from M. jannaschii | | Descriptor: | Dihydroorotase, ZINC ION | | Authors: | Vitali, J, Nix, J.C, Newman, H.E, Colaneri, M.J. | | Deposit date: | 2022-04-12 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Methanococcus jannaschii dihydroorotase.
Proteins, 91, 2023
|
|
7WW2
 
 | | Structure of an Isocytosine specific deaminase Vcz | | Descriptor: | 8-oxoguanine deaminase, ZINC ION | | Authors: | Li, X.J, Wu, B.X. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of an isocytosine-specific deaminase VCZ reveals its application potential in the anti-cancer therapy.
Iscience, 26, 2023
|
|
7X68
 
 | | CYS179 and CYS504 of CRMP2 were covalently binded by a Sesquiterpene lactone | | Descriptor: | (3aR,5S,8R,8aR,9aR)-5,8a-dimethyl-3-methylidene-8-oxidanyl-5,6,7,8,9,9a-hexahydro-3aH-benzo[f][1]benzofuran-2-one, Dihydropyrimidinase-related protein 2, SODIUM ION | | Authors: | Zhang, S.D, Ma, Y.F, Zhang, J. | | Deposit date: | 2022-03-06 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CYS179 and CYS504 of CRMP2 were covalently binded by a Sesquiterpene lactone
To Be Published
|
|
4RDV
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-FORMIMINO-L-GLUTAMATE IMINOHYDROLASE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate
To be Published
|
|
4RZB
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate, SOAKED WITH MERCURY | | Descriptor: | GLYCEROL, MERCURY (II) ION, N-[(E)-iminomethyl]-L-aspartic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-12-19 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
4TQT
 
 | |
5LP3
 
 | |
5MLE
 
 | | Crystal Structure of Human Dihydropyrimidinease-like 2 (DPYSL2A)/Collapsin Response Mediator Protein (CRMP2 13-516) Mutant Y479E/Y499E | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Dihydropyrimidinase-related protein 2, ... | | Authors: | Sethi, R, Zheng, Y, Talon, R, Velupillai, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ahmed, A.A, von Delft, F. | | Deposit date: | 2016-12-06 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Tuning microtubule dynamics to enhance cancer therapy by modulating FER-mediated CRMP2 phosphorylation.
Nat Commun, 9, 2018
|
|
5LXX
 
 | | High-resolution structure of human collapsin response mediator protein 2 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dihydropyrimidinase-related protein 2, SULFATE ION | | Authors: | Myllykoski, M, Hensley, K, Kursula, P. | | Deposit date: | 2016-09-23 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Collapsin response mediator protein 2: high-resolution crystal structure sheds light on small-molecule binding, post-translational modifications, and conformational flexibility.
Amino Acids, 49, 2017
|
|
5NKS
 
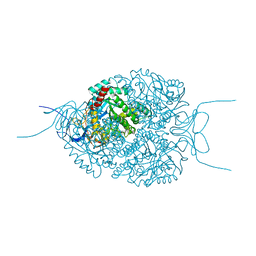 | | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4) | | Descriptor: | Dihydropyrimidinase-related protein 4 | | Authors: | Mathea, S, Elkins, J.M, Strain-Damerell, C, Salah, E, Borkowska, O, Chalk, R, Burgess-Brown, N, Pinkas, D.M, von Delft, F, Krojer, T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2017-03-31 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Dihydropyrimidinase-related Protein 4 (DPYSL4, CRMP3, ULIP-4)
To Be Published
|
|
5MKV
 
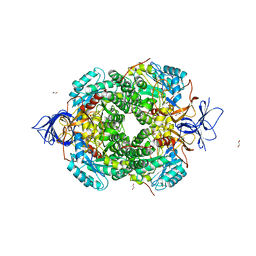 | | Crystal Structure of Human Dihydropyrimidinease-like 2 (DPYSL2A)/Collapsin Response Mediator Protein (CRMP2) residues 13-516 | | Descriptor: | 1,2-ETHANEDIOL, Dihydropyrimidinase-related protein 2 | | Authors: | Sethi, R, Zheng, Y, Krojer, T, Velupillai, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ahmed, A.A, von Delft, F. | | Deposit date: | 2016-12-05 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tuning microtubule dynamics to enhance cancer therapy by modulating FER-mediated CRMP2 phosphorylation.
Nat Commun, 9, 2018
|
|
3SFW
 
 | |
