2XIU
 
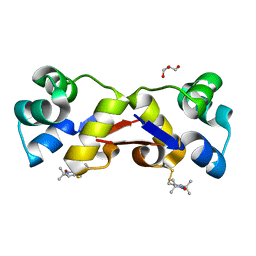 | | High resolution structure of MTSL-tagged CylR2. | | Descriptor: | CYLR2, GLYCEROL, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XJ3
 
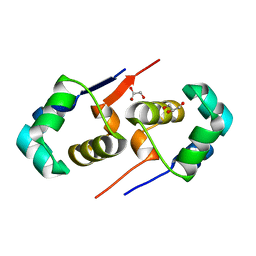 | | High resolution structure of the T55C mutant of CylR2. | | Descriptor: | CYLR2 SYNONYM CYTOLYSIN REPRESSOR 2, GLYCEROL | | Authors: | Gruene, T, Cho, M.K, Karyagina, I, Kim, H.Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-02 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2L49
 
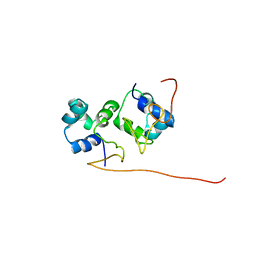 | | The solution structure of the P2 C,the immunity repressor of the P2 bacteriophage | | Descriptor: | C protein | | Authors: | Massad, T, Papadopolos, E, Stenmark, P, Damberg, P. | | Deposit date: | 2010-10-01 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The C repressor of the P2 bacteriophage.
J.Biomol.Nmr, 64, 2016
|
|
3OP9
 
 | |
2XCJ
 
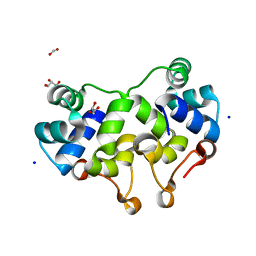 | | Crystal structure of P2 C, the immunity repressor of temperate E. coli phage P2 | | Descriptor: | C PROTEIN, FORMIC ACID, GLYCEROL, ... | | Authors: | Massad, T, Skaar, K, Hogbom, M, Stenmark, P. | | Deposit date: | 2010-04-23 | | Release date: | 2010-07-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the P2 C-Repressor: A Binder of Non-Palindromic Direct DNA Repeats.
Nucleic Acids Res., 38, 2010
|
|
3KZ3
 
 | |
3KXA
 
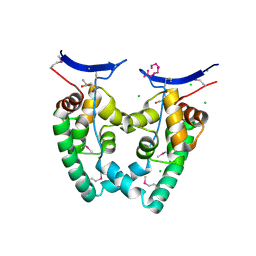 | | Crystal Structure of NGO0477 from Neisseria gonorrhoeae | | Descriptor: | ASPARAGINE, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Ren, J, Sainsbury, S, Nettleship, J.E, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-12-02 | | Release date: | 2010-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of NGO0477 from Neisseria gonorrhoeae reveals a novel protein fold incorporating a helix-turn-helix motif.
Proteins, 78, 2010
|
|
3JXC
 
 | |
3JXB
 
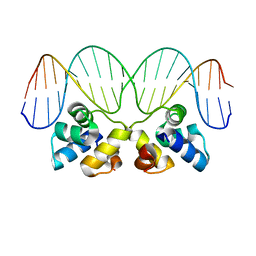 | |
3JXD
 
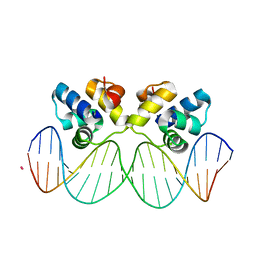 | |
2KPJ
 
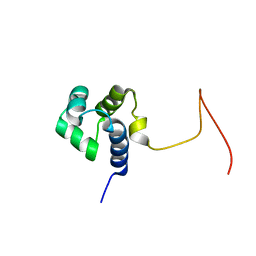 | | Solution Structure Of Protein SOS-response transcriptional repressor, LexA From Eubacterium rectale. Northeast Structural Genomics Consortium Target ErR9A | | Descriptor: | SOS-response transcriptional repressor, LexA | | Authors: | Wu, Y, Eletsky, A, Lee, D, Ghosh, A, Buchwald, W, Zhang, Q, Janjua, H, Garcia, E, Nair, R, Sukumaran, D, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-16 | | Release date: | 2009-11-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein SOS-response transcriptional repressor, LexA From Eubacterium rectale. Northeast Structural Genomics Consortium Target ErR9A
To be Published
|
|
3IVP
 
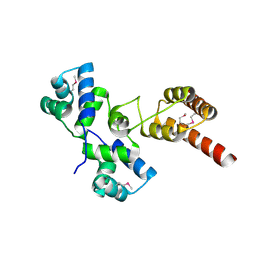 | |
3FYA
 
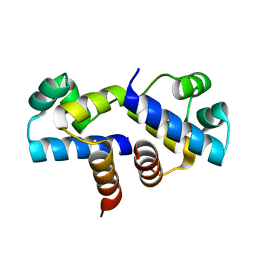 | | Crystal Structure of an R35A mutant of the Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | Regulatory protein | | Authors: | Ball, N.J, McGeehan, J.E, Thresh, S.J, Streeter, S.D, Kneale, G.G. | | Deposit date: | 2009-01-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the restriction-modification controller protein C.Esp1396I.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3G5G
 
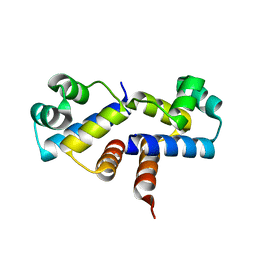 | | Crystal Structure of the Wild-Type Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | Regulatory protein | | Authors: | Ball, N.J, McGeehan, J.E, Thresh, S.J, Streeter, S.D, Kneale, G.G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the restriction-modification controller protein C.Esp1396I.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3HZI
 
 | | Structure of mdt protein | | Descriptor: | 5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3', ADENOSINE-5'-TRIPHOSPHATE, HTH-type transcriptional regulator hipB, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
2WIU
 
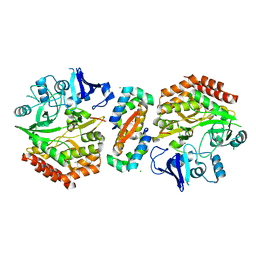 | | Mercury-modified bacterial persistence regulator hipBA | | Descriptor: | CHLORIDE ION, HTH-TYPE TRANSCRIPTIONAL REGULATOR HIPB, MERCURY (II) ION, ... | | Authors: | Evdokimov, A, Voznesensky, I, Fennell, K, Anderson, M, Smith, J.F, Fisher, D.A. | | Deposit date: | 2009-05-17 | | Release date: | 2009-07-28 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New Kinase Regulation Mechanism Found in Hipba: A Bacterial Persistence Switch.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3DNV
 
 | | MDT Protein | | Descriptor: | DNA (5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3'), HTH-type transcriptional regulator hipB, Protein hipA, ... | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3EUS
 
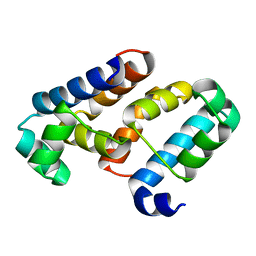 | |
2JVL
 
 | | NMR structure of the C-terminal domain of MBF1 of Trichoderma reesei | | Descriptor: | TrMBF1 | | Authors: | Kopke Salinas, R, Tomaselli, S, Camilo, C.M, Valencia, E.Y, Farah, C.S, El-Dorry, H, Chambergo, F.S. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of multiprotein bridging factor 1 (MBF1) of Trichoderma reesei.
Proteins, 75, 2009
|
|
3CLC
 
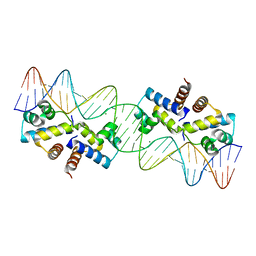 | | Crystal Structure of the Restriction-Modification Controller Protein C.Esp1396I Tetramer in Complex with its Natural 35 Base-Pair Operator | | Descriptor: | 35-MER, MAGNESIUM ION, Regulatory protein | | Authors: | McGeehan, J.E, Streeter, S.D, Thresh, S.J, Ball, N, Ravelli, R.B, Kneale, G.G. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of the genetic switch that regulates the expression of restriction-modification genes.
Nucleic Acids Res., 36, 2008
|
|
2R1J
 
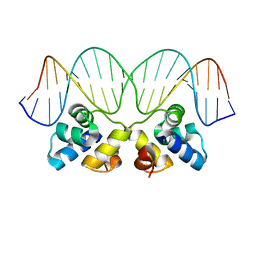 | | Crystal Structure of the P22 c2 Repressor protein in complex with the synthetic operator 9T | | Descriptor: | 5'-D(*DCP*DAP*DTP*DTP*DTP*DAP*DAP*DGP*DAP*DTP*DAP*DTP*DCP*DTP*DTP*DAP*DAP*DAP*DTP*DA)-3', 5'-D(*DTP*DAP*DTP*DTP*DTP*DAP*DAP*DGP*DAP*DTP*DAP*DTP*DCP*DTP*DTP*DAP*DAP*DAP*DTP*DG)-3', Repressor protein C2 | | Authors: | Williams, L.D, Koudelka, G.B, Watkins, D, Hsiao, C, Woods, K. | | Deposit date: | 2007-08-22 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | P22 c2 repressor-operator complex: mechanisms of direct and indirect readout
Biochemistry, 47, 2008
|
|
3BDN
 
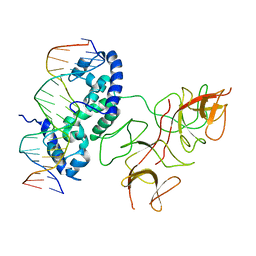 | | Crystal Structure of the Lambda Repressor | | Descriptor: | DNA (5'-D(*DAP*DAP*DTP*DAP*DCP*DCP*DAP*DCP*DTP*DGP*DGP*DCP*DGP*DGP*DTP*DGP*DAP*DTP*DAP*DT)-3'), DNA (5'-D(*DTP*DAP*DTP*DAP*DTP*DCP*DAP*DCP*DCP*DGP*DCP*DCP*DAP*DGP*DTP*DGP*DGP*DTP*DAP*DT)-3'), Lambda Repressor | | Authors: | Stayrook, S.E, Jaru-Ampornpan, P, Hochschild, A, Lewis, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.909 Å) | | Cite: | Crystal structure of the lambda repressor and a model for pairwise cooperative operator binding
Nature, 452, 2008
|
|
3CEC
 
 | |
2EF8
 
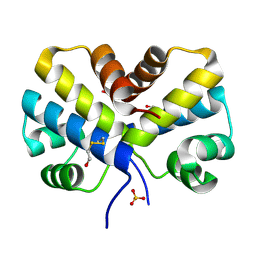 | |
3BS3
 
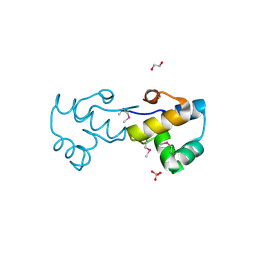 | | Crystal structure of a putative DNA-binding protein from Bacteroides fragilis | | Descriptor: | 1,2-ETHANEDIOL, Putative DNA-binding protein, SULFATE ION | | Authors: | Cuff, M.E, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of a putative DNA-binding protein from Bacteroides fragilis.
TO BE PUBLISHED
|
|
