6EHS
 
 | | E. coli Hydrogenase-2 chemically reduced structure | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, DITHIONITE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Evans, R.M, Beaton, S.E, Armstrong, F.A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
6EN9
 
 | | E. coli Hydrogenase-2 (hydrogen reduced form) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
6FPI
 
 | | Structure of fully reduced Hydrogenase (Hyd-1) variant E28Q | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6FPO
 
 | | High resolution structure of native Hydrogenase (Hyd-1) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6FPW
 
 | | Structure of fully reduced Hydrogenase (Hyd-1) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6CFW
 
 | |
6G2J
 
 | | Mouse mitochondrial complex I in the active state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Agip, A.N.A, Blaza, J.N, Bridges, H.R, Viscomi, C, Rawson, S, Muench, S.P, Hirst, J. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-06 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6G72
 
 | | Mouse mitochondrial complex I in the deactive state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Acyl carrier protein, mitochondrial, ... | | Authors: | Agip, A.N.A, Blaza, J.N, Bridges, H.R, Viscomi, C, Rawson, S, Muench, S.P, Hirst, J. | | Deposit date: | 2018-04-04 | | Release date: | 2018-06-06 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of complex I from mouse heart mitochondria in two biochemically defined states.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6G7M
 
 | | Four-site variant (Y222C, C197S, C432S, C433S) of E. coli hydrogenase-2 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Zhang, L, Beaton, S.E. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Direct visible light activation of a surface cysteine-engineered [NiFe]-hydrogenase by silver nanoclusters
Energy Environ Sci, 2019
|
|
6G7R
 
 | | Structure of fully reduced variant E28Q of E. coli hydrogenase-1 at pH 8 | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6G94
 
 | |
6GAL
 
 | | Structure of fully reduced Hydrogenase (Hyd-1) variant E28Q collected at pH 10 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6GAM
 
 | | Structure of E14Q variant of E. coli hydrogenase-2 (as-isolated enzyme) | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6GAN
 
 | | Structure of fully reduced Hydrogenase (Hyd-2) variant E14Q | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Evans, R.M. | | Deposit date: | 2018-04-11 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic Exploitation of a Self-Repairing, Blocked Proton Transfer Pathway in an O2-Tolerant [NiFe]-Hydrogenase.
J. Am. Chem. Soc., 140, 2018
|
|
6GCS
 
 | |
6H8K
 
 | | Crystal structure of a variant (Q133C in PSST) of Yarrowia lipolytica complex I | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, NADH dehydrogenase [ubiquinone] flavoprotein 1, ... | | Authors: | Wirth, C, Galemou Yoga, E, Zickermann, V, Hunte, C. | | Deposit date: | 2018-08-02 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Locking loop movement in the ubiquinone pocket of complex I disengages the proton pumps.
Nat Commun, 9, 2018
|
|
6HUM
 
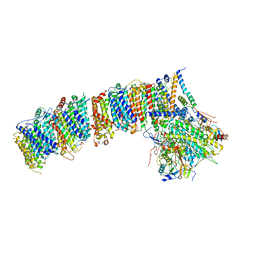 | | Structure of the photosynthetic complex I from Thermosynechococcus elongatus | | Descriptor: | 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, IRON/SULFUR CLUSTER, ... | | Authors: | Schuller, J.M, Schuller, S.K, Kurisu, G, Engel, B.D, Nowaczyk, M.M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer.
Science, 363, 2019
|
|
6I0D
 
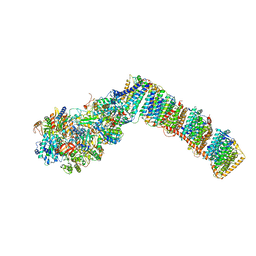 | | Respiratory complex I from Thermus thermophilus with bound Decyl-Ubiquinone | | Descriptor: | 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-10-25 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6I1P
 
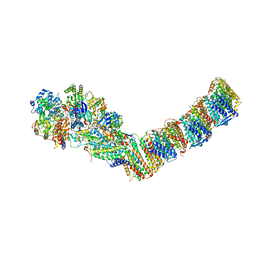 | | Respiratory complex I from Thermus thermophilus with bound NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-10-29 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.207 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6NBQ
 
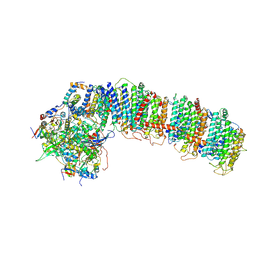 | | T.elongatus NDH (data-set 1) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 2, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-09 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBX
 
 | | T.elongatus NDH (data-set 2) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBY
 
 | | T.elongatus NDH (composite model) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2020-04-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6Q8O
 
 | | Respiratory complex I from Thermus thermophilus with bound Piericidin A | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-12-15 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.605 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6Q8X
 
 | | Respiratory complex I from Thermus thermophilus with bound Pyridaben. | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-12-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6Q8W
 
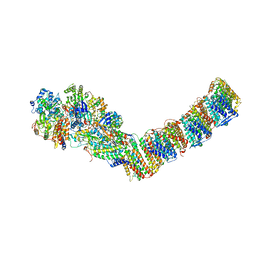 | | Respiratory complex I from Thermus thermophilus with bound Aureothin. | | Descriptor: | Aureothin, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2018-12-16 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
