3V9Z
 
 | |
3V9S
 
 | |
3V9U
 
 | |
3VA3
 
 | |
6A46
 
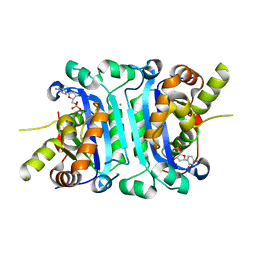 | | Structure of TREX2 in complex with a nucleotide (dCMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hsiao, Y.Y. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the duplex DNA processing of TREX2
Nucleic Acids Res., 46, 2018
|
|
6A4B
 
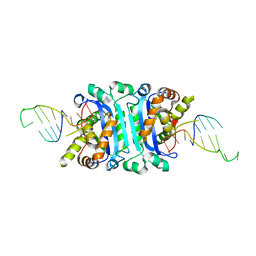 | |
3VA0
 
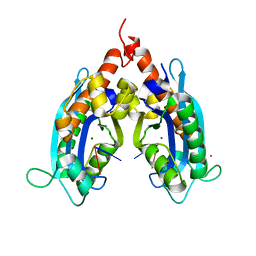 | |
3V9X
 
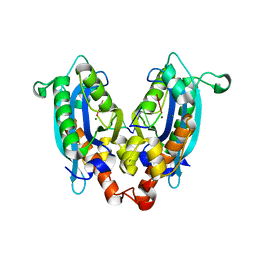 | |
6A47
 
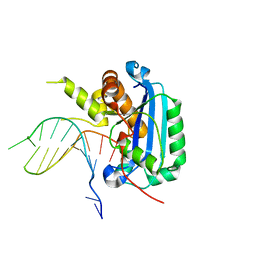 | | Structure of TREX2 in complex with a Y structured dsDNA | | Descriptor: | DNA (5'-D(P*CP*CP*AP*GP*GP*CP*CP*CP*TP*CP*TP*AP*GP*GP*GP*CP*CP*TP*T)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Hsiao, Y.Y. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the duplex DNA processing of TREX2
Nucleic Acids Res., 46, 2018
|
|
6A4D
 
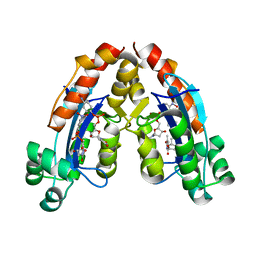 | |
6A4E
 
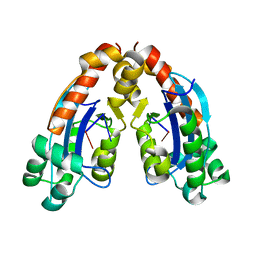 | |
6A4A
 
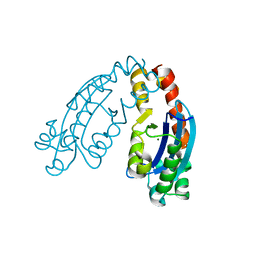 | |
6A4F
 
 | |
6A45
 
 | | Structure of mouse TREX2 | | Descriptor: | PHOSPHATE ION, Three prime repair exonuclease 2 | | Authors: | Hsiao, Y.Y. | | Deposit date: | 2018-06-19 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural insights into the duplex DNA processing of TREX2
Nucleic Acids Res., 46, 2018
|
|
3V9W
 
 | |
6N6D
 
 | | Vibrio cholerae Oligoribonuclease bound to pAG | | Descriptor: | Oligoribonuclease, RNA (5'-R(P*AP*G)-3'), SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6K
 
 | | Human REXO2 bound to pAG | | Descriptor: | MALONATE ION, RNA (5'-R(P*AP*G)-3'), RNA exonuclease 2 homolog,Small fragment nuclease, ... | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6J
 
 | | Human REXO2 bound to pAA | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MALONATE ION, ... | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.317 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6F
 
 | | Vibrio cholerae Oligoribonuclease bound to pGC | | Descriptor: | Oligoribonuclease, RNA (5'-R(P*GP*C)-3'), SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6E
 
 | | Vibrio cholerae Oligoribonuclease bound to pGA | | Descriptor: | RNA (5'-R(P*GP*A)-3'), RNA exonuclease 2 homolog,Small fragment nuclease, SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6A
 
 | | Vibrio cholerae Oligoribonuclease bound to pGG | | Descriptor: | Oligoribonuclease, RNA (5'-R(P*GP*G)-3'), SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6G
 
 | | Vibrio cholerae Oligoribonuclease bound to pCG | | Descriptor: | Oligoribonuclease, RNA (5'-R(P*CP*G)-3'), SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6C
 
 | | Vibrio cholerae Oligoribonuclease bound to pAA | | Descriptor: | RNA (5'-R(P*AP*A)-3'), RNA exonuclease 2 homolog,Small fragment nuclease, SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6H
 
 | | Vibrio cholerae Oligoribonuclease bound to pCpU | | Descriptor: | Oligoribonuclease, RNA (5'-R(P*CP*U)-3'), SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.757 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6I
 
 | | Human REXO2 bound to pGG | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MALONATE ION, ... | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.431 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
