7ANB
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7ANA
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7AN1
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1636-S1_20) | | Descriptor: | Arylsulfatase, CALCIUM ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-10 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7ALL
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT4683-S1_4) | | Descriptor: | Arylsulfatase, CALCIUM ION, IODIDE ION, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Martens, E.C, Basle, A, Cartmell, A. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
6XLP
 
 | | Structure of the essential inner membrane lipopolysaccharide-PbgA complex | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-deoxy-3-O-[(1R,3R)-1,3-dihydroxytetradecyl]-2-{[(3R)-3-hydroxytetradecanoyl]amino}-1-O-phosphono-alpha-D-glucopyranose-(6-1)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)]1,5-anhydro-2-deoxy-2-{[(1S,3R)-1-hydroxy-3-(pentanoyloxy)undecyl]amino}-4-O-phosphono-D-glucitol, ... | | Authors: | Payandeh, J, Clairefeuille, T. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the essential inner membrane lipopolysaccharide-PbgA complex.
Nature, 584, 2020
|
|
6VDF
 
 | |
6VC7
 
 | |
6VAT
 
 | |
6V8Q
 
 | | Structure of an inner membrane protein required for PhoPQ regulated increases in outer membrane cardiolipin | | Descriptor: | (9Z,21R,24R,30R,33R,44Z)-24,27,30-trihydroxy-18,24,30,36-tetraoxo-19,23,25,29,31,35-hexaoxa-24lambda~5~,30lambda~5~-dip hosphatripentaconta-9,44-diene-21,33-diyl (9Z,9'Z)di-octadec-9-enoate, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fan, J, Miller, S. | | Deposit date: | 2019-12-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.699585 Å) | | Cite: | Structure of an Inner Membrane Protein Required for PhoPQ-Regulated Increases in Outer Membrane Cardiolipin.
Mbio, 11, 2020
|
|
6UST
 
 | | Gut microbial sulfatase from Hungatella hathewayi | | Descriptor: | CALCIUM ION, N-acetylgalactosamine 6-sulfate sulfatase | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Endobiotic Reactivation by Human Gut Microbiome-Encoded Sulfatases.
Biochemistry, 59, 2020
|
|
6USS
 
 | |
6SUT
 
 | | Crystal structure of phosphothreonine MCR-2 | | Descriptor: | BROMIDE ION, GLYCEROL, Putative integral membrane protein, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-09-16 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Resistance to the "last resort" antibiotic colistin: a single-zinc mechanism for phosphointermediate formation in MCR enzymes.
Chem.Commun.(Camb.), 56, 2020
|
|
6S21
 
 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33494S-sulf) | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Endo-4-O-sulfatase | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
6S20
 
 | | Metabolism of multiple glycosaminoglycans by bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus (BT33336S-sulf) | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, N-acetylgalactosamine-6-O-sulfatase, ... | | Authors: | Ndeh, D, Basle, A, Strahl, H, Henrissat, B, Terrapon, N, Cartmell, A. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Metabolism of multiple glycosaminoglycans by Bacteroides thetaiotaomicron is orchestrated by a versatile core genetic locus.
Nat Commun, 11, 2020
|
|
6LI6
 
 | | Crystal structure of MCR-1-S treated by Au(PEt3)Cl | | Descriptor: | GOLD ION, Probable phosphatidylethanolamine transferase Mcr-1, TRIETHYLPHOSPHANE | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LI5
 
 | | Crystal structure of apo-MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1 | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LI4
 
 | | Crystal structure of MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6J66
 
 | | Chondroitin sulfate/dermatan sulfate endolytic 4-O-sulfatase | | Descriptor: | CALCIUM ION, Chondroitin sulfate/dermatan sulfate 4-O-endosulfatase protein | | Authors: | Gu, L, Li, F, Su, T, Wang, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Comparative Study of Two Chondroitin Sulfate/Dermatan Sulfate 4-O-Sulfatases With High Identity.
Front Microbiol, 10, 2019
|
|
6IOZ
 
 | | Structural insights of idursulfase beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kim, H, Kim, D, Hong, J, Lee, K, Seo, J, Oh, B.H. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights of idursulfase beta
To Be Published
|
|
6HR5
 
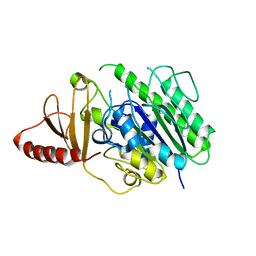 | | Structure of the S1_25 family sulfatase module of the rhamnosidase FA22250 from Formosa agariphila | | Descriptor: | Alpha-L-rhamnosidase/sulfatase (GH78), CALCIUM ION | | Authors: | Roret, T, Prechoux, A, Czjzek, M, Michel, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.912 Å) | | Cite: | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
6HHM
 
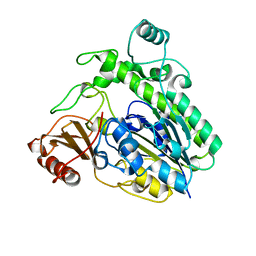 | | Crystal structure of the family S1_7 ulvan-specific sulfatase FA22070 from Formosa agariphila | | Descriptor: | Arylsulfatase, CALCIUM ION | | Authors: | Roret, T, Prechoux, A, Michel, G, Czjzek, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
6G60
 
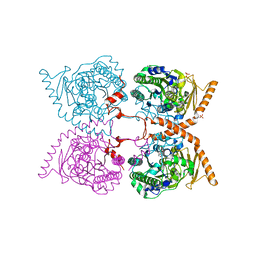 | |
6G5Z
 
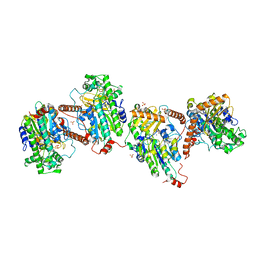 | |
6FNY
 
 | | CRYSTAL STRUCTURE OF A CHOLINE SULFATASE FROM SINORHIZOBIUM MELLILOTI | | Descriptor: | CALCIUM ION, Choline-sulfatase | | Authors: | Valkov, E, Van Loo, B, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and Mechanistic Analysis of the Choline Sulfatase from Sinorhizobium melliloti: A Class I Sulfatase Specific for an Alkyl Sulfate Ester.
J. Mol. Biol., 430, 2018
|
|
6DGM
 
 | | Streptococcus pyogenes phosphoglycerol transferase GacH in complex with sn-glycerol-1-phosphate | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Phosphoglycerol transferase GacH, ... | | Authors: | Edgar, R.J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-05-17 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Discovery of glycerol phosphate modification on streptococcal rhamnose polysaccharides.
Nat.Chem.Biol., 15, 2019
|
|
