5VNP
 
 | |
5W8P
 
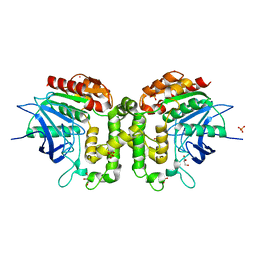 | | Homoserine transacetylase MetX from Mycobacterium abscessus | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase, PHOSPHATE ION, ... | | Authors: | Rodriguez, E.S, Reed, R.W, Korotkov, K.V. | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of mycobacterial homoserine transacetylases central to methionine biosynthesis reveals druggable active site.
Sci Rep, 9, 2019
|
|
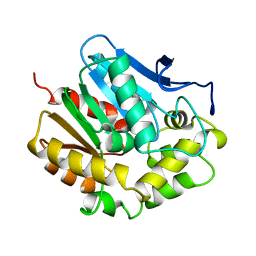
2HAD
 
 | |
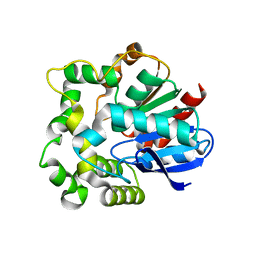
4NZZ
 
 | |
6COB
 
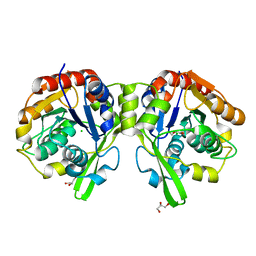 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,F179L,A209I | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
6COI
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,A209I with CYANIDE, benzaldehyde, MANDELIC ACID NITRILE | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, (S)-MANDELIC ACID NITRILE, Alpha-hydroxynitrile lyase, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.024 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
6CL4
 
 | | LipC12 - Lipase from metagenomics | | Descriptor: | Lipase C12 | | Authors: | Iulek, J, Martini, V.P, Krieger, N, Glogauer, A, Souza, E.M. | | Deposit date: | 2018-03-01 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure solution and analyses of the first true lipase obtained from metagenomics indicate potential for increased thermostability.
N Biotechnol, 53, 2019
|
|
6COG
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,A209I with benzaldehyde | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
5K3E
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Glycolate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
6COH
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,A209I with benzaldehyde, MANDELIC ACID NITRILE | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, Alpha-hydroxynitrile lyase, GLYCEROL, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
6COC
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,F179L,A209I with benzaldehyde | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
6COF
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo, Y13C,Y121L,P126F,L128W,C131T,A209I | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
5JZS
 
 | | HsaD bound to 3,5-dichloro-4-hydroxybenzoic acid | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD, 3,5-dichloro-4-hydroxybenzoic acid | | Authors: | Ryan, A, Polycarpou, E, Lack, N, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E.D, Ballet, R, Abuhammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5K3A
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Both Protomers Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
6COD
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo Y13C,Y121L,P126F,L128W,C131T,F179L,A209I with benzaldehyde | | Descriptor: | Alpha-hydroxynitrile lyase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
6COE
 
 | | AtHNL enantioselectivity mutant At-A9-H7 Apo Y13C,Y121L,P126F,L128W,C131T,F179L,A209I with benzaldehyde, MANDELIC ACID NITRILE | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, (S)-MANDELIC ACID NITRILE, Alpha-hydroxynitrile lyase, ... | | Authors: | Jones, B.J, Kazlauskas, R.J, Desrouleaux, R. | | Deposit date: | 2018-03-12 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | AtHNL enantioselectivity mutants
To be published
|
|
5K3B
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Chloroacetate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, chloroacetic acid | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5JZB
 
 | | Crystal structure of HsaD bound to 3,5-dichlorobenzene sulphonamide | | Descriptor: | 3,5-dichlorobenzene-1-sulfonamide, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, PHOSPHATE ION | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5K3C
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - WT/5-Fluorotryptophan | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5K3D
 
 | |
5JZ9
 
 | | Crystal structure of HsaD bound to 3,5-dichloro-4-hydroxybenzenesulphonic acid | | Descriptor: | 3,5-dichloro-4-hydroxybenzene-1-sulfonic acid, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5K3F
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Single Protomer Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
4OSE
 
 | |
5LKA
 
 | | Crystal structure of haloalkane dehalogenase LinB 140A+143L+177W+211L mutant (LinB86) from Sphingobium japonicum UT26 at 1.3 A resolution | | Descriptor: | Haloalkane dehalogenase, THIOCYANATE ION | | Authors: | Degtjarik, O, Rezacova, P, Iermak, I, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2016-07-21 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Engineering a de novo transport tunnel.
Acs Catalysis, 2016
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
