3M4F
 
 | |
3LB9
 
 | | Crystal structure of the B. circulans cpA123 circular permutant | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
3EXU
 
 | | A glycoside hydrolase family 11 xylanase with an extended thumb region | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase, GLYCEROL | | Authors: | Vandermarliere, E, Pollet, A, Strelkov, S.V, Delcour, J.A, Courtin, C.M. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic and activity-based evidence for thumb flexibility and its relevance in glycoside hydrolase family 11 xylanases
Proteins, 77, 2009
|
|
3HD8
 
 | | Crystal structure of the Triticum aestivum xylanase inhibitor-IIA in complex with bacillus subtilis xylanase | | Descriptor: | Endo-1,4-beta-xylanase A, Xylanase inhibitor | | Authors: | Sansen, S, Pollet, A, Raedschelders, G, Gebruers, K, Rabijns, A, Courtin, C.M. | | Deposit date: | 2009-05-07 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Identification of structural determinants for inhibition strength and specificity of wheat xylanase inhibitors TAXI-IA and TAXI-IIA
Febs J., 276, 2009
|
|
2VUL
 
 | | Thermostable mutant of ENVIRONMENTALLY ISOLATED GH11 XYLANASE | | Descriptor: | DODECAETHYLENE GLYCOL, GH11 XYLANASE, SULFATE ION | | Authors: | Dumon, C, Varvak, A, Wall, M.A, Flint, J.E, Lewis, R.J, Lakey, J.H, Luginbuhl, P, Healey, S, Todaro, T, Desantis, G, Sun, M, Parra-Gessert, L, Tan, X, Weiner, D.P, Gilbert, H.J. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering Hyperthermostability Into a Gh11 Xylanase is Mediated by Subtle Changes to Protein Structure.
J.Biol.Chem., 283, 2008
|
|
2JIC
 
 | | High resolution structure of xylanase-II from one micron beam experiment | | Descriptor: | XYLANASE-II | | Authors: | Moukhametzianov, R, Burghammer, M, Edwards, P.C, Petitdemange, S, Popov, D, Fransen, M, Schertler, G.F, Riekel, C. | | Deposit date: | 2007-02-27 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Crystallography with a Micrometre-Sized Synchrotron-Radiation Beam.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3B5L
 
 | |
2VG9
 
 | | Crystal structure of Loop Swap mutant of Necallimastix patriciarum Xyn11A | | Descriptor: | ACETATE ION, BIFUNCTIONAL ENDO-1,4-BETA-XYLANASE A, CADMIUM ION | | Authors: | Vardakou, M, Dumon, C, Flint, J.E, Murray, J.W, Christakopoulos, P, Weiner, D.P, Juge, N, Lewis, R.J, Gilbert, H.J. | | Deposit date: | 2007-11-09 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the Structural Basis for Substrate and Inhibitor Recognition in Eukaryotic Gh11 Xylanases.
J.Mol.Biol., 375, 2008
|
|
2QZ2
 
 | | Crystal structure of a glycoside hydrolase family 11 xylanase from Aspergillus niger in complex with xylopentaose | | Descriptor: | Endo-1,4-beta-xylanase I, SODIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Vandermarliere, E, Rombouts, S, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
2Z79
 
 | | High resolution crystal structure of a glycoside hydrolase family 11 xylanase of Bacillus subtilis | | Descriptor: | Endo-1,4-beta-xylanase A, GLYCEROL | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
2QZ3
 
 | | Crystal structure of a glycoside hydrolase family 11 xylanase from Bacillus subtilis in complex with xylotetraose | | Descriptor: | ACETIC ACID, Endo-1,4-beta-xylanase A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
2NQY
 
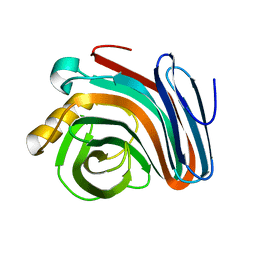 | |
2DCJ
 
 | | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
2DCK
 
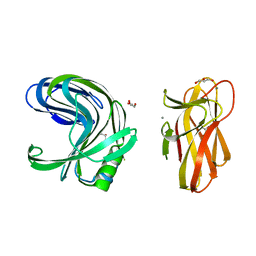 | | A tetragonal-lattice structure of alkaliphilic XynJ from Bacillus sp. 41M-1 | | Descriptor: | CALCIUM ION, GLYCEROL, xylanase J | | Authors: | Fibriansah, G, Ihsanawati, Tanaka, N, Nakamura, S, Kumasaka, T. | | Deposit date: | 2006-01-07 | | Release date: | 2007-03-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1
To be Published
|
|
2C1F
 
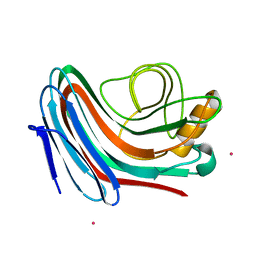 | |
2B42
 
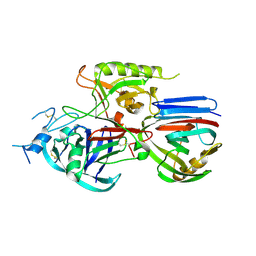 | | Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase | | Descriptor: | Endo-1,4-beta-xylanase A, xylanase inhibitor-I | | Authors: | Sansen, S, Dewilde, M, De Ranter, C.J, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-09-19 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of structural determinants for inhibition strength and specificity of wheat xylanase inhibitors TAXI-IA and TAXI-IIA.
Febs J., 276, 2009
|
|
2B45
 
 | | Crystal structure of an engineered uninhibited Bacillus subtilis xylanase in free state | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endo-1,4-beta-xylanase A | | Authors: | Sansen, S, Dewilde, M, De Ranter, C.J, Sorensen, J.F, Sibbesen, O, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase
To be Published
|
|
2B46
 
 | | Crystal structure of an engineered uninhibited Bacillus subtilis xylanase in substrate bound state | | Descriptor: | Endo-1,4-beta-xylanase A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Sansen, S, Dewilde, M, De Ranter, C.J, Sorensen, J.F, Sibbesen, O, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2005-09-22 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase
To be Published
|
|
2D98
 
 | | Structure of VIL (extra KI/I2 added)-xylanase | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-09 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2D97
 
 | | Structure of VIL-xylanase | | Descriptor: | Endo-1,4-beta-xylanase 2 | | Authors: | Miyatake, H, Hasegawa, T, Yamano, A. | | Deposit date: | 2005-12-09 | | Release date: | 2006-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | New methods to prepare iodinated derivatives by vaporizing iodine labelling (VIL) and hydrogen peroxide VIL (HYPER-VIL)
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DFC
 
 | | Xylanase II from Tricoderma reesei at 293K | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Harata, K, Akiba, T. | | Deposit date: | 2006-02-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DFB
 
 | | Xylanase II from Tricoderma reesei at 100K | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, SULFATE ION | | Authors: | Harata, K, Akiba, T. | | Deposit date: | 2006-02-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure of an orthorhombic form of xylanase II from Trichoderma reesei and analysis of thermal displacement.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
2DCZ
 
 | | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase By Directed Evolution | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Endo-1,4-beta-xylanase A, SULFATE ION | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2DCY
 
 | | Crystal structure of Bacillus subtilis family-11 xylanase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D(-)-TARTARIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Kondo, H, Miyazaki, K, Takenouchi, M, Noro, N, Suzuki, M, Tsuda, S. | | Deposit date: | 2006-01-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Thermal Stabilization of Bacillus subtilis Family-11 Xylanase by Directed Evolution
J.Biol.Chem., 281, 2006
|
|
2F6B
 
 | |
