4EGY
 
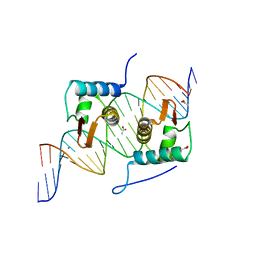 | | Crystal Structure of AraR(DBD) in complex with operator ORA1 | | Descriptor: | 5'-D(*AP*AP*AP*AP*TP*TP*GP*TP*TP*CP*GP*TP*AP*CP*AP*AP*AP*TP*AP*TP*T)-3', 5'-D(*TP*AP*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*AP*AP*CP*AP*AP*TP*TP*T)-3', ACETATE ION, ... | | Authors: | Jain, D, Nair, D.T. | | Deposit date: | 2012-04-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators.
Nucleic Acids Res., 41, 2013
|
|
4EGZ
 
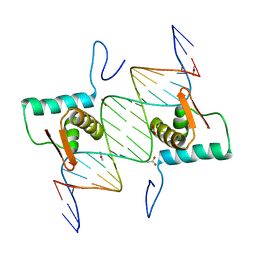 | | Crystal Structure of AraR(DBD) in complex with operator ORR3 | | Descriptor: | 5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*CP*CP*GP*TP*AP*TP*AP*CP*AP*TP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*AP*TP*GP*TP*AP*TP*AP*CP*GP*GP*AP*CP*AP*AP*AP*TP*T)-3', ACETATE ION, ... | | Authors: | Jain, D, Nair, D.T. | | Deposit date: | 2012-04-02 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators.
Nucleic Acids Res., 41, 2013
|
|
4H0E
 
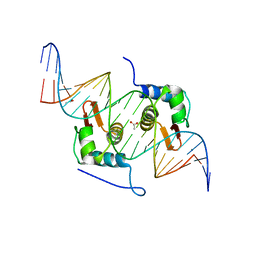 | | Crystal Structure of mutant ORR3 in complex with NTD of AraR | | Descriptor: | 5'-D(*AP*AP*AP*TP*TP*TP*GP*TP*CP*CP*GP*TP*AP*CP*AP*TP*TP*TP*TP*AP*T)-3', 5'-D(*TP*AP*TP*AP*AP*AP*AP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*AP*AP*TP*T)-3', ACETATE ION, ... | | Authors: | Nair, D.T, Jain, D. | | Deposit date: | 2012-09-08 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Spacing between core recognition motifs determines relative orientation of AraR monomers on bipartite operators.
Nucleic Acids Res., 41, 2013
|
|
4MGR
 
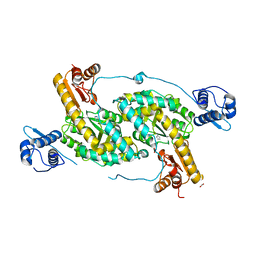 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4N0B
 
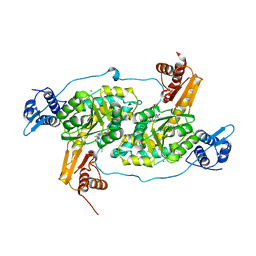 | | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of GabT | | Descriptor: | ACETYL GROUP, CALCIUM ION, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Edayathumangalam, R, Wu, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-30 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4R1H
 
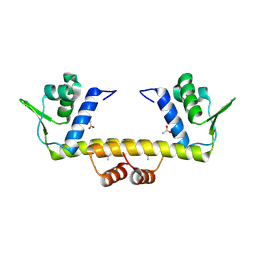 | |
4TV7
 
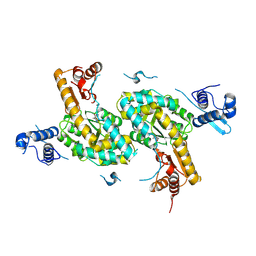 | |
4WWC
 
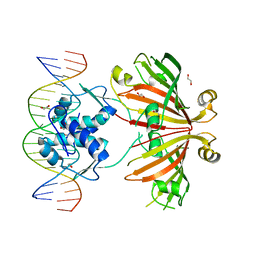 | | Crystal structure of full length YvoA in complex with palindromic operator DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*AP*GP*TP*GP*GP*TP*CP*TP*AP*GP*AP*CP*CP*AP*CP*TP*GP*G)-3'), HTH-type transcriptional repressor YvoA | | Authors: | Grau, F.C, Fillenberg, S.B, Muller, Y.A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural insight into operator dre-sites recognition and effector binding in the GntR/HutC transcription regulator NagR.
Nucleic Acids Res., 43, 2015
|
|
4U0V
 
 | |
4U0W
 
 | |
4U0Y
 
 | |
4P96
 
 | |
4P9U
 
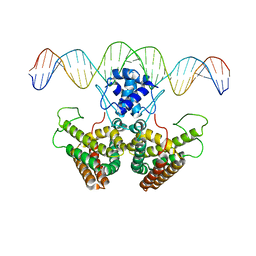 | |
4PDK
 
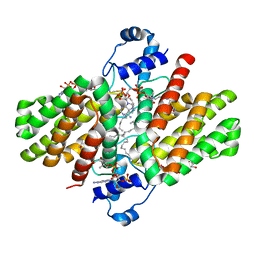 | | FadR, Fatty Acid Responsive Transcription Factor from Vibrio cholerae, in Complex with oleoyl-CoA | | Descriptor: | Fatty acid metabolism regulator protein, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name) | | Authors: | Shi, W, Kull, F.J. | | Deposit date: | 2014-04-19 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 40-residue insertion in Vibrio cholerae FadR facilitates binding of an additional fatty acyl-CoA ligand.
Nat Commun, 6, 2015
|
|
5D4S
 
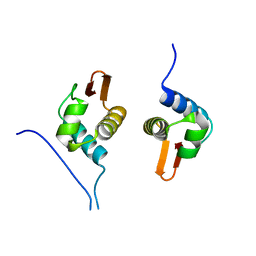 | | Crystal Structure of AraR(DBD) in complex with operator ORX1 | | Descriptor: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*AP*AP*TP*AP*CP*AP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*TP*GP*TP*AP*TP*T)-3') | | Authors: | Jain, D, Narayanan, N, Nair, D.T. | | Deposit date: | 2015-08-08 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
5D4R
 
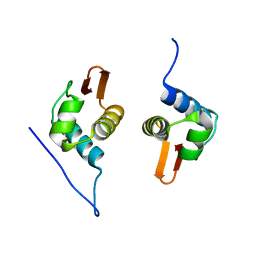 | | Crystal Structure of AraR(DBD) in complex with operator ORE1 | | Descriptor: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*CP*TP*AP*AP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*TP*AP*GP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*A)-3') | | Authors: | Jain, D, Narayanan, N, Nair, D.T. | | Deposit date: | 2015-08-08 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
4ZS8
 
 | | Crystal structure of ligand-free, full length DasR | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional repressor DasR | | Authors: | Fillenberg, S.B, Muller, Y.A. | | Deposit date: | 2015-05-13 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the Global Regulator DasR from Streptomyces coelicolor: Implications for the Allosteric Regulation of GntR/HutC Repressors.
Plos One, 11, 2016
|
|
5KVR
 
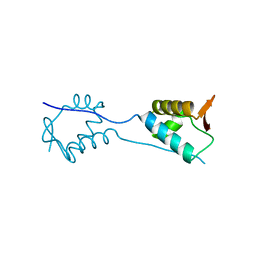 | | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073 | | Descriptor: | Pyruvate dehydrogenase complex repressor | | Authors: | Brunzelle, J.S, Wawrzak, Z, Sandoval, J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | X-Ray Crystal Structure of a Fragment (1-75) of a Transcriptional Regulator PdhR from Escherichia coli CFT073
To Be Published
|
|
5XGF
 
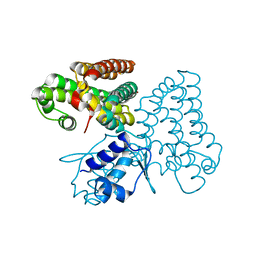 | |
6AZ6
 
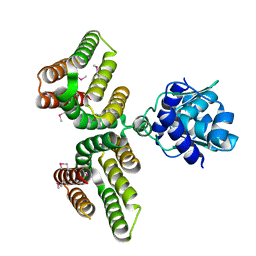 | | Streptococcus agalactiae GntR | | Descriptor: | GntR family transcriptional regulator | | Authors: | Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2017-09-10 | | Release date: | 2017-12-20 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Structural basis for the regulation of beta-glucuronidase expression by human gut Enterobacteriaceae.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EP3
 
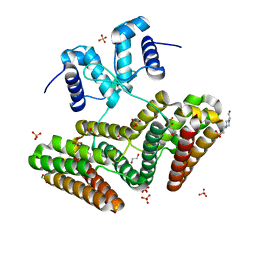 | | Lar controls the expression of the Listeria monocytogenes agr system and mediates virulence. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lmo0651 protein, ... | | Authors: | Pinheiro, J, Lisboa, J, Carvalho, F, Carreaux, A, Santos, N, Cabral, J, Sousa, S, Cabanes, D. | | Deposit date: | 2017-10-10 | | Release date: | 2018-07-25 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MouR controls the expression of the Listeria monocytogenes Agr system and mediates virulence.
Nucleic Acids Res., 46, 2018
|
|
6SBS
 
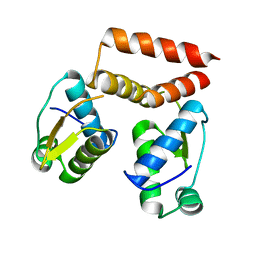 | | YtrA from Sulfolobus acidocaldarius, a GntR-family transcription factor | | Descriptor: | Regulatory protein | | Authors: | Lemmens, L, Valegard, K, Lindas, A.C, Peeters, E, Maes, D. | | Deposit date: | 2019-07-22 | | Release date: | 2019-07-31 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | YtrASa, a GntR-Family Transcription Factor, Represses Two Genetic Loci Encoding Membrane Proteins inSulfolobus acidocaldarius.
Front Microbiol, 10, 2019
|
|
6ON4
 
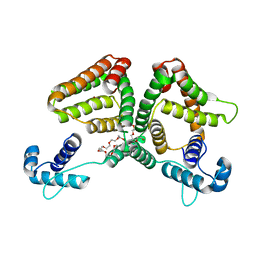 | | Crystal structure of the GntR-type sialoregulator NanR from Escherichia coli, in complex with sialic acid | | Descriptor: | HTH-type transcriptional repressor NanR, N-acetyl-beta-neuraminic acid, ZINC ION, ... | | Authors: | Horne, C.R, Panjikar, S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2019-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Control of the Escherichia coli sialoregulon by transcriptional repressor NanR.
J. Bacteriol., 195, 2013
|
|
6Z74
 
 | |
6ZA7
 
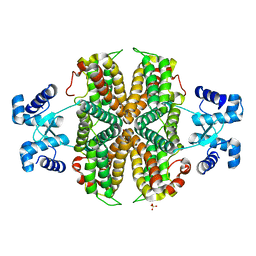 | | Structure of the apo transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
