6JVO
 
 | | Crystal structure of human MTH1 in complex with compound MI1022 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVM
 
 | | Crystal structure of human MTH1 in complex with compound MI1016 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-5-ethyl-6-piperidin-1-yl-pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVF
 
 | | Crystal structure of human apo MTH1 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Peng, C, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
6JVL
 
 | | Crystal structure of human MTH1 in complex with compound MI1014 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N4-cyclopropyl-5-ethyl-6-(4-methylpiperazin-1-yl)pyrimidine-2,4-diamine | | Authors: | Peng, C, Li, Y.H, Cheng, Y.S. | | Deposit date: | 2019-04-17 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitor development of MTH1 via high-throughput screening with fragment based library and MTH1 substrate binding cavity.
Bioorg.Chem., 110, 2021
|
|
5ANW
 
 | | MTH1 in complex with compound 24 | | Descriptor: | 2-[4-(2-AMINOQUINAZOLIN-4-YL)PHENYL]-N,N-DIMETHYL-ACETAMIDE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5ANT
 
 | | Potent and selective inhibitors of MTH1 probe its role in cancer cell survival | | Descriptor: | 2-(2-methoxyethoxy)-6-(methylamino)-9-(phenylmethyl)-7H-purin-8-one, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Kettle, J.G, Alwan, H, Bista, M, Breed, J, Kack, H, Eckersley, K, Foote, K.M, Fillery, S, Goodwin, L, Jones, D, Lau, A, Nissink, J.W.M, Read, J, Scott, J, Taylor, B, Walker, G, Wissler, L. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5BON
 
 | | Crystal structure of human Nudt15 (MTH2) | | Descriptor: | MAGNESIUM ION, Probable 8-oxo-dGTP diphosphatase NUDT15 | | Authors: | Carter, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | Deposit date: | 2015-05-27 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2.
Nat Commun, 6, 2015
|
|
5ANS
 
 | | Potent and selective inhibitors of MTH1 probe its role in cancer cell survival | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Kettle, J.G, Alwan, H, Bista, M, Breed, J, Kack, H, Eckersley, K, Foote, K.M, Fillery, S, Goodwin, L, Jones, D, Lau, A, Nissink, J.W.M, Read, J, Scott, J, Taylor, B, Walker, G, Wissler, L. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5ANV
 
 | | MTH1 in complex with compound 15 | | Descriptor: | 4-(4-CHLORO-2-FLUORO-ANILINO)-6,7-DIMETHOXY-N-METHYL-QUINOLINE-3-CARBOXAMIDE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5ANU
 
 | | MTH1 in complex with compound 15 | | Descriptor: | 13-(METHYLAMINO)-23,24,25-TRIOXA-17,18,19,21-TETRAZATETRACYCLO-TRICOSA-1(3),2(10),4(11),12(14),13(18),16(19)-HEXAN-15-ONE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, DIMETHYL SULFOXIDE | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2015-09-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potent and Selective Inhibitors of Mth1 Probe its Role in Cancer Cell Survival.
J.Med.Chem., 59, 2016
|
|
5C8L
 
 | | Crystal Structure of the Bdellovibrio bacteriovorus Nucleoside Diphosphate Sugar Hydrolase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NudF protein, ... | | Authors: | Gabelli, S.B, de la Pena, A.H, Suarez, A, Amzel, L.M. | | Deposit date: | 2015-06-25 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus.
Plos One, 10, 2015
|
|
5C7Q
 
 | | Crystal Structure of the Bdellovibrio bacteriovorus Nucleoside Diphosphate Sugar Hydrolase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Gabelli, S.B, de la Pena, A.H, Suarez, A, Amzel, L.M. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus.
Plos One, 10, 2015
|
|
5CFJ
 
 | | Structural and functional attributes of malaria parasite Ap4A hydrolase | | Descriptor: | BIS(5'-nucleosyl)-tetraphosphatase (Diadenosine tetraphosphatase), putative, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sharma, A, Yogavel, M, Sharma, A. | | Deposit date: | 2015-07-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and functional attributes of malaria parasite diadenosine tetraphosphate hydrolase
Sci Rep, 6, 2016
|
|
5C7T
 
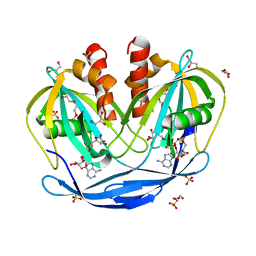 | | Crystal Structure of the Bdellovibrio bacteriovorus Nucleoside Diphosphate Sugar Hydrolase in complex with ADP-ribose | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ADENOSINE-5-DIPHOSPHORIBOSE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gabelli, S.B, de la Pena, A.H, Suarez, A, Amzel, L.M. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and Enzymatic Characterization of a Nucleoside Diphosphate Sugar Hydrolase from Bdellovibrio bacteriovorus.
Plos One, 10, 2015
|
|
5CFI
 
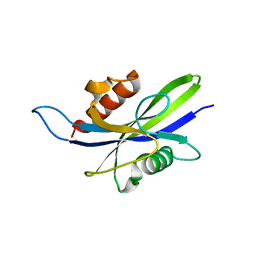 | |
6NL1
 
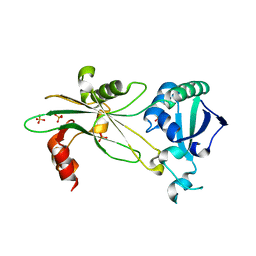 | | Structure of T. brucei MERS1 protein in its apo form | | Descriptor: | Mitochondrial edited mRNA stability factor 1, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
1K26
 
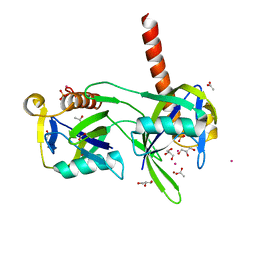 | | Structure of a Nudix Protein from Pyrobaculum aerophilum Solved by the Single Wavelength Anomolous Scattering Method | | Descriptor: | ACETIC ACID, GLYCEROL, IRIDIUM (III) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1JKN
 
 | | Solution Structure of the Nudix Enzyme Diadenosine Tetraphosphate Hydrolase from Lupinus angustifolius Complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, diadenosine 5',5'''-P1,P4-tetraphosphate hydrolase | | Authors: | Fletcher, J.I, Swarbrick, J.D, Maksel, D, Gayler, K.R, Gooley, P.R. | | Deposit date: | 2001-07-12 | | Release date: | 2002-02-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The structure of Ap(4)A hydrolase complexed with ATP-MgF(x) reveals the basis of substrate binding.
Structure, 10, 2002
|
|
1K2E
 
 | | crystal structure of a nudix protein from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-09-26 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1VIU
 
 | |
1VIQ
 
 | |
3COU
 
 | | Crystal structure of human Nudix motif 16 (NUDT16) | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 16 | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nyman, T, Persson, C, Sagemark, J, Schueler, H, Svensson, L, Thorsell, A.G, Van Den Berg, S, Welin, M, Weigelt, J, Wikstrom, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-29 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human Nudix motif 16 (NUDT16).
To be Published
|
|
1VHZ
 
 | | Crystal structure of ADP compounds hydrolase | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP compounds hydrolase nudE | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHG
 
 | | Crystal structure of ADP compounds hydrolase | | Descriptor: | ADP compounds hydrolase nudE | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1JRK
 
 | | Crystal Structure of a Nudix Protein from Pyrobaculum aerophilum Reveals a Dimer with Intertwined Beta Sheets | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nudix homolog | | Authors: | Wang, S, Mura, C, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2001-08-13 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Nudix protein from Pyrobaculum aerophilum reveals a dimer with two intersubunit beta-sheets.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
