2JG2
 
 | | HIGH RESOLUTION STRUCTURE OF SPT WITH PLP INTERNAL ALDIMINE | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-05-01 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
5AX8
 
 | | Recombinant expression, purification and preliminary crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, mitochondrial | | Authors: | Jiang, X, Wang, J, Chang, H, Zhou, Y. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Recombinant expression, purification and crystallographic studies of the mature form of human mitochondrial aspartate aminotransferase
Biosci Trends, 10, 2016
|
|
3MEB
 
 | |
5BK7
 
 | | The structure of MppP E15A mutant soaked with the substrate L-arginine | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-27 | | Release date: | 2018-09-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
5BJ4
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 | | Descriptor: | PHOSPHATE ION, PROTEIN (ASPARTATE AMINOTRANSFERASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Kawaguchi, S.I, Miyahara, I, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-01-11 | | Release date: | 2003-09-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme
J.BIOCHEM.(TOKYO), 130, 2001
|
|
2GB3
 
 | |
2JGT
 
 | | Low resolution structure of SPT | | Descriptor: | SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-14 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
7V5I
 
 | |
7V58
 
 | | Structural insights into the substrate selectivity of acyl-CoA transferase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-amino-3-ketobutyrate coenzyme A ligase, GLYCINE, ... | | Authors: | Chang, H.Y, Ko, T.P. | | Deposit date: | 2021-08-16 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insights into the substrate selectivity of alpha-oxoamine synthases from marine Vibrio sp. QWI-06.
Colloids Surf B Biointerfaces, 210, 2022
|
|
5EAA
 
 | | ASPARTATE AMINOTRANSFERASE FROM E. COLI, C191S MUTATION | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-12-29 | | Release date: | 2000-10-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase.
Protein Eng., 13, 2000
|
|
7TUR
 
 | | Joint X-ray/neutron structure of aspastate aminotransferase (AAT) in complex with pyridoxamine 5'-phosphate (PMP) | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-2-METHYL-SUCCINIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aspartate aminotransferase, ... | | Authors: | Drago, V.N, Kovalevsky, A.Y, Dajnowicz, S, Mueser, T.C. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | An N⋯H⋯N low-barrier hydrogen bond preorganizes the catalytic site of aspartate aminotransferase to facilitate the second half-reaction.
Chem Sci, 13, 2022
|
|
5DJ3
 
 | | Structure of the PLP-Dependent L-Arginine Hydroxylase MppP with D-Arginine Bound | | Descriptor: | (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-D-arginine, MAGNESIUM ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.227 Å) | | Cite: | Streptomyces wadayamensis MppP Is a Pyridoxal 5'-Phosphate-Dependent l-Arginine alpha-Deaminase, gamma-Hydroxylase in the Enduracididine Biosynthetic Pathway.
Biochemistry, 54, 2015
|
|
5DJ1
 
 | | Structure of the PLP-Dependent L-Arginine Hydroxylase MppP Holoenzyme | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2015-09-01 | | Release date: | 2015-11-25 | | Last modified: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Streptomyces wadayamensis MppP Is a Pyridoxal 5'-Phosphate-Dependent l-Arginine alpha-Deaminase, gamma-Hydroxylase in the Enduracididine Biosynthetic Pathway.
Biochemistry, 54, 2015
|
|
5EFS
 
 | | The crystal structure of human kynurenine aminotransferase II | | Descriptor: | Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Nematollahi, A, Sun, G, Kwan, A, Harrop, S.J, Hanrahan, J.R, Nadvi, N.A, Church, W.B. | | Deposit date: | 2015-10-26 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82503 Å) | | Cite: | The crystal structure of human kynurenine aminotransferase II
To Be Published
|
|
7U7H
 
 | | Cysteate acyl-ACP transferase from Alistipes finegoldii | | Descriptor: | 7-keto-8-aminopelargonate synthetase-like enzyme, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Radka, C.D, Miller, D.J, Frank, M.W, Rock, C.O. | | Deposit date: | 2022-03-07 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biochemical characterization of the first step in sulfonolipid biosynthesis in Alistipes finegoldii.
J.Biol.Chem., 298, 2022
|
|
7VRX
 
 | | Pad-1 in the absence of substrate | | Descriptor: | Aminotransferase, SULFATE ION | | Authors: | Choi, M, Rhee, S. | | Deposit date: | 2021-10-25 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96634674 Å) | | Cite: | Structural and biochemical basis for the substrate specificity of Pad-1, an indole-3-pyruvic acid aminotransferase in auxin homeostasis.
J.Struct.Biol., 214, 2022
|
|
5EUN
 
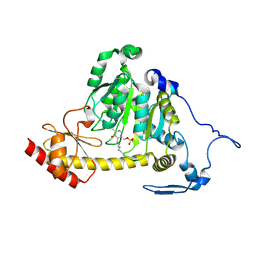 | | The Crystal Structure of Human Kynurenine Aminotransferase II, PLP-bound form, at 1.83 A | | Descriptor: | Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Nematollahi, A, Sun, G, Kwan, A, Jeffries, C.M, Harrop, S.J, Hanrahan, J.R, Nadvi, N.A, Church, W.B. | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-16 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Structure of the PLP-Form of the Human Kynurenine Aminotransferase II in a Novel Spacegroup at 1.83 angstrom Resolution.
Int J Mol Sci, 17, 2016
|
|
2Q7W
 
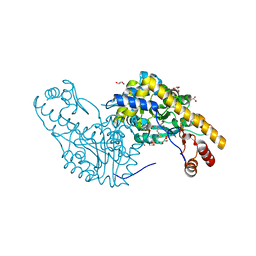 | | Structural Studies Reveals the Inactivation of E. coli L-aspartate aminotransferase (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via two mechanisms at pH 6.0 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inactivation of Escherichia coli l-Aspartate Aminotransferase by (S)-4-Amino-4,5-dihydro-2-thiophenecarboxylic Acid Reveals "A Tale of Two Mechanisms".
Biochemistry, 46, 2007
|
|
2QB2
 
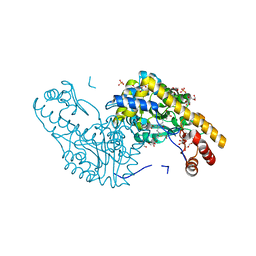 | | Structural Studies Reveal the Inactivation of E. coli L-aspartate aminotransferase by (s)-4,5-dihydro-2thiophenecarboylic acid (SADTA) via two mechanisms (at pH 7.0). | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
6DND
 
 | | Crystal structure of wild-type (WT) human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
2QLR
 
 | |
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6DNB
 
 | | Crystal structure of T110A:S256A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, GLYCEROL, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
5VK7
 
 | |
