8APH
 
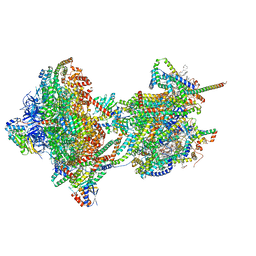 | | rotational state 2c of the Trypanosoma brucei mitochondrial ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8AP6
 
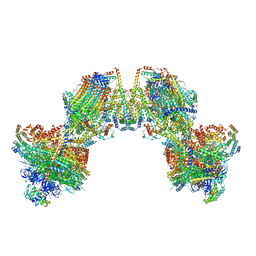 | | Trypanosoma brucei mitochondrial F1Fo ATP synthase dimer | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Muehleip, A, Gahura, O, Zikova, A, Amunts, A. | | Deposit date: | 2022-08-09 | | Release date: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | An ancestral interaction module promotes oligomerization in divergent mitochondrial ATP synthases.
Nat Commun, 13, 2022
|
|
8EAT
 
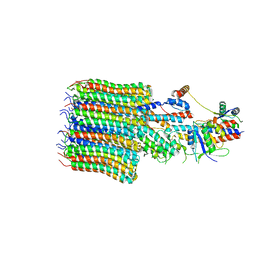 | | Yeast VO missing subunits a, e, and f in complex with Vma12-22p | | Descriptor: | V-type proton ATPase subunit F, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAU
 
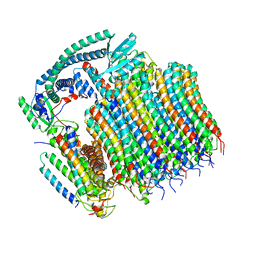 | | Yeast VO in complex with Vma21p | | Descriptor: | V-type proton ATPase subunit a, vacuolar isoform, V-type proton ATPase subunit c, ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EAS
 
 | | Yeast VO in complex with Vma12-22p | | Descriptor: | V-type proton ATPase assembly factor Vma12p, V-type proton ATPase subunit F, V-type proton ATPase subunit a, ... | | Authors: | Wang, H, Bueler, S.A, Rubinstein, J.L. | | Deposit date: | 2022-08-29 | | Release date: | 2022-11-02 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of V-ATPase V O region assembly by Vma12p, 21p, and 22p.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H9U
 
 | | Human ATP synthase state 3a (combined) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9M
 
 | | Human ATP synthase state 3a subregion 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9J
 
 | | Human ATP synthase state2 subregion 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9V
 
 | | Human ATP synthase state 3b (combined) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9T
 
 | | Human ATP synthase state 2 (combined) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9Q
 
 | | Human ATP synthase state 3b subregion 3 | | Descriptor: | ATP synthase F(0) complex subunit B1, mitochondrial, ATP synthase F(0) complex subunit C1, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9S
 
 | | Human ATP synthase state 1 (combined) | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-05-31 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8H9F
 
 | | Human ATP synthase state 1 subregion 3 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
8F29
 
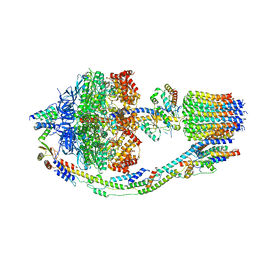 | | Yeast ATP synthase in conformation-1 at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8F39
 
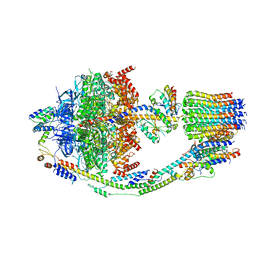 | | Yeast ATP synthase in conformation-2, at pH 6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase protein 8, ATP synthase subunit 4, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-11-09 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FKJ
 
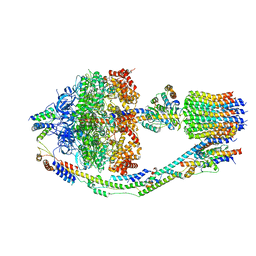 | | Yeast ATP Synthase in conformation-3, at pH 6 | | Descriptor: | ATP synthase protein 8, ATP synthase subunit 4, mitochondrial, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FL8
 
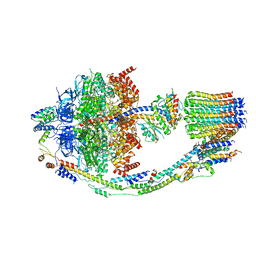 | | Yeast ATP Synthase structure in presence of MgATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase protein 8, ... | | Authors: | Sharma, S, Patel, H, Luo, M, Mueller, D.M, Liao, M. | | Deposit date: | 2022-12-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Conformational ensemble of yeast ATP synthase at low pH reveals unique intermediates and plasticity in F 1 -F o coupling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8G0E
 
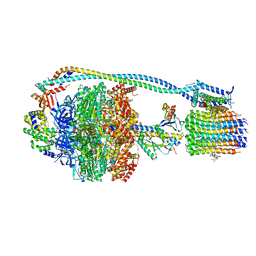 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0C
 
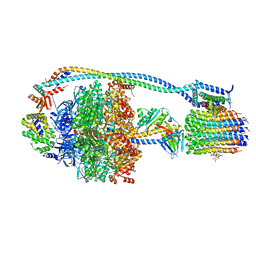 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0B
 
 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase FO region | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ATP synthase subunit a, ATP synthase subunit b, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G09
 
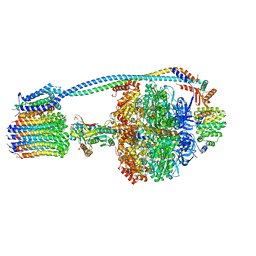 | |
8G07
 
 | |
8G08
 
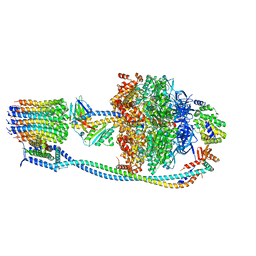 | |
8G0D
 
 | | Cryo-EM structure of TBAJ-876-bound Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | (1R,2S)-1-(6-bromo-2-methoxyquinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2,3,6-trimethoxypyridin-4-yl)butan-2-ol, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2023-01-31 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of mycobacterial ATP synthase inhibition by squaramides and second generation diarylquinolines.
Embo J., 42, 2023
|
|
8G0A
 
 | |
