2BZ7
 
 | |
2IDF
 
 | | P. aeruginosa azurin N42C/M64E double mutant, BMME-linked dimer | | Descriptor: | 1-[PYRROL-1-YL-2,5-DIONE-METHOXYMETHYL]-PYRROLE-2,5-DIONE, Azurin, COPPER (II) ION, ... | | Authors: | Einsle, O, de Jongh, T.E, Hoffmann, M, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-09-15 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electron transfer in a crosslinked protein dimer mediated by a hydrogen-bonded network across the dimer interface
To be Published
|
|
2IWE
 
 | | Structure of a cavity mutant (H117G) of Pseudomonas aeruginosa azurin | | Descriptor: | 1,1'-HEXANE-1,6-DIYLBIS(1H-IMIDAZOLE), AZURIN, ZINC ION | | Authors: | De Jongh, T.E, Van Roon, A.M.M, Prudencio, M, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-06-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Click-Chemistry with an Active Site Variant of Azurin
Eur.J.Inorg.Chem., 2006, 2006
|
|
2Q5B
 
 | | High resolution structure of Plastocyanin from Phormidium laminosum | | Descriptor: | ACETATE ION, COPPER (II) ION, POTASSIUM ION, ... | | Authors: | Fromme, R, Bukhman-DeRuyter, Y.S, Grotjohann, I, Mi, H, Fromme, P. | | Deposit date: | 2007-05-31 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Plastocyanin at high resolution from Phormidium laminosum and the double mutant D44A, D45A
To be Published
|
|
2QDV
 
 | | Structure of the Cu(II) form of the M51A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION | | Authors: | Carrell, C.J, Ma, J.K, Wang, Y, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2007-06-21 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Biochemistry, 46, 2007
|
|
2QDW
 
 | | Structure of Cu(I) form of the M51A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION, PHOSPHATE ION | | Authors: | Ma, J.K, Wang, Y, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2007-06-21 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Biochemistry, 46, 2007
|
|
2JKW
 
 | | Pseudoazurin M16F | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Yanagisawa, S, Crowley, P.B, Firbank, S.J, Lawler, A.T, Hunter, D.M, McFarlane, W, Li, C, Kohzuma, T, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pi-Interaction Tuning of the Active Site Properties of Metalloproteins.
J.Am.Chem.Soc., 130, 2008
|
|
2RAC
 
 | | AMICYANIN REDUCED, PH 7.7, 1.3 ANGSTROMS | | Descriptor: | COPPER (I) ION, PROTEIN (AMICYANIN) | | Authors: | Cunane, L.M, Chen, Z.-W, Durley, R.C.E, Mathews, F.S. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for interprotein complex-dependent effects on the redox properties of amicyanin.
Biochemistry, 37, 1998
|
|
2TSB
 
 | | AZURIN MUTANT M121A-AZIDE | | Descriptor: | AZIDE ION, AZURIN AZIDE, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2TSA
 
 | | AZURIN MUTANT M121A | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2UX7
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of loop shortening on the metal binding site of cupredoxin pseudoazurin.
Biochemistry, 46, 2007
|
|
2UXG
 
 | | Pseudoazurin with engineered amicyanin ligand loop, reduced form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2UXF
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 5.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2UX6
 
 | | Pseudoazurin with engineered amicyanin ligand loop, oxidized form, pH 7.5 | | Descriptor: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Velarde, M, Huber, R, Yanagisawa, S, Dennison, C, Messerschmidt, A. | | Deposit date: | 2007-03-27 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of Loop Shortening on the Metal Binding Site of Cupredoxin Pseudoazurin.
Biochemistry, 46, 2007
|
|
2W88
 
 | |
2W8C
 
 | |
2N0M
 
 | | The solution structure of the soluble form of the Lipid-modified Azurin from Neisseria gonorrhoeae | | Descriptor: | COPPER (I) ION, Lipid modified azurin protein | | Authors: | Pauleta, S.R, Matzapetakis, M.F, Nobrega, C.F, Carreira, C, Saraiva, I.H. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the soluble form of the lipid-modified azurin from Neisseria gonorrhoeae, the electron donor of cytochrome c peroxidase.
Biochim.Biophys.Acta, 1857, 2016
|
|
2XV2
 
 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.2 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
2OJ1
 
 | | Disulfide-linked dimer of azurin N42C/M64E double mutant | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | de Jongh, T.E, Hoffmann, M, Einsle, O, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2007-01-12 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inter- and intramolecular electron transfer in modified azurin dimers
Eur.J.Inorg.Chem., 2007, 2007
|
|
2PCY
 
 | |
2OV0
 
 | | Structure of the blue copper protein Amicyanin to 0.75 A resolution | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Carrell, C.J, Davidson, V.L, Chen, Z, Cunane, L.M, Trickey, P, Mathews, F.S. | | Deposit date: | 2007-02-12 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Ultrahigh resolution studies of amicyanin
TO BE PUBLISHED
|
|
2PLT
 
 | |
1QHQ
 
 | | AURACYANIN, A BLUE COPPER PROTEIN FROM THE GREEN THERMOPHILIC PHOTOSYNTHETIC BACTERIUM CHLOROFLEXUS AURANTIACUS | | Descriptor: | CHLORIDE ION, COPPER (II) ION, PROTEIN (AURACYANIN), ... | | Authors: | Bond, C.S, Blankenship, R.E, Freeman, H.C, Guss, J.M, Maher, M, Selvaraj, F, Wilce, M.C.J, Willingham, K. | | Deposit date: | 1999-05-25 | | Release date: | 2001-03-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of auracyanin, a "blue" copper protein from the green thermophilic photosynthetic bacterium Chloroflexus aurantiacus.
J.Mol.Biol., 306, 2001
|
|
4DP1
 
 | | The 1.35 Angstrom crystal structure of reduced (CuI) poplar plastocyanin B at pH 4.0 | | Descriptor: | ACETATE ION, COPPER (I) ION, GLYCEROL, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
4DP7
 
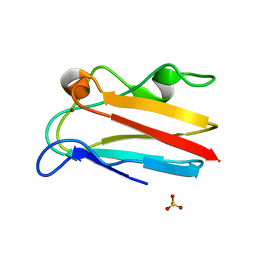 | | The 1.08 Angstrom crystal structure of oxidized (CuII) poplar plastocyanin A at pH 4.0 | | Descriptor: | COPPER (II) ION, Plastocyanin A, chloroplastic, ... | | Authors: | Kachalova, G.S, Shosheva, A.H, Bourenkov, G.P, Donchev, A.A, Dimitrov, M.I, Bartunik, H.D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Structural comparison of the poplar plastocyanin isoforms PCa and PCb sheds new light on the role of the copper site geometry in interactions with redox partners in oxygenic photosynthesis.
J.Inorg.Biochem., 115, 2012
|
|
