2Q9J
 
 | | Crystal structure of the C217S mutant of diaminopimelate epimerase | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
6PG0
 
 | | Protein Tyrosine Phosphatase 1B (1-301), P188A mutant, vanadate bound state | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, VANADATE ION | | Authors: | Cui, D.S, Lipchock, J.M, Loria, J.P. | | Deposit date: | 2019-06-23 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
2PI2
 
 | | Full-length Replication protein A subunits RPA14 and RPA32 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Borgstahl, G.E. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Full-length Human RPA14/32 Complex Gives Insights into the Mechanism of DNA Binding and Complex Formation.
J.Mol.Biol., 374, 2007
|
|
6TG0
 
 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 21a | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, SULFATE ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
7QPP
 
 | | High resolution structure of human VDR ligand binding domain in complex with calcitriol | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, SULFATE ION, Vitamin D3 receptor | | Authors: | Rochel, N. | | Deposit date: | 2022-01-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Advances in Vitamin D Receptor Function and Evolution Based on the 3D Structure of the Lamprey Ligand-Binding Domain.
J.Med.Chem., 65, 2022
|
|
7QRD
 
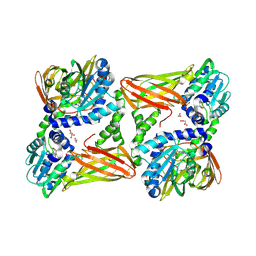 | | Crystal structure of mouse CARM1 in complex with histone H3_10-25 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-ETHANEDIOL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2022-01-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARM1 Transition State Mimics
To Be Published
|
|
9HGT
 
 | | Amyloid fibril from the antimicrobial peptide citropin 1-3 - Polymorph Nr.1L | | Descriptor: | Citropin-1.3 | | Authors: | Strati, F, Landau, M, Siavash, M, Cali, P.M, Monistrol, J, Golubev, A, Gustavsson, E, Bloch, Y. | | Deposit date: | 2024-11-20 | | Release date: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of citropin 1.3 amyloid polymorph
To Be Published
|
|
6VJ7
 
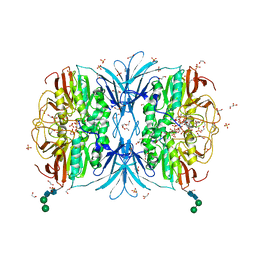 | | Crystal structure of red kidney bean purple acid phosphatase in complex with adenosine 5'-(beta,gamma imido)triphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Schenk, G, Guddat, L.W, McGeary, R.P, Mitic, N, Furtado, A, Schulz, B.L, Henry, R.J, Schmidt, S. | | Deposit date: | 2020-01-15 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elements that modulate the substrate specificity of plant purple acid phosphatases: Avenues for improved phosphorus acquisition in crops.
Plant Sci., 294, 2020
|
|
7A6T
 
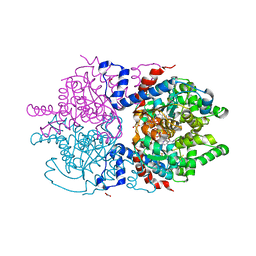 | | Crystal Structure of Asn173Ser variant of Human Deoxyhypusine Synthase in complex with NAD and spermidine | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Deoxyhypusine synthase, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-08-26 | | Release date: | 2022-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
2PN9
 
 | | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA modified aptamer | | Descriptor: | 5'-R(*GP*GP*AP*GP*CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*C)-3', RNA 16-mer with locked residues 9-10 | | Authors: | Lebars, I, Richard, T, Di Primo, C, Toulme, J.-J. | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer
Nucleic Acids Res., 35, 2007
|
|
3KKU
 
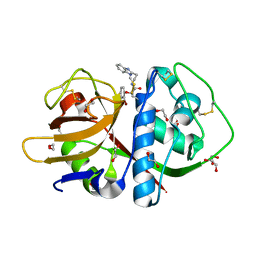 | | Cruzain in complex with a non-covalent ligand | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, N-[2-(1H-benzimidazol-2-yl)ethyl]-2-(2-bromophenoxy)acetamide, ... | | Authors: | Ferreira, R.S, Eidam, O, Shoichet, B.K. | | Deposit date: | 2009-11-06 | | Release date: | 2010-07-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Complementarity between a docking and a high-throughput screen in discovering new cruzain inhibitors.
J.Med.Chem., 53, 2010
|
|
3KN6
 
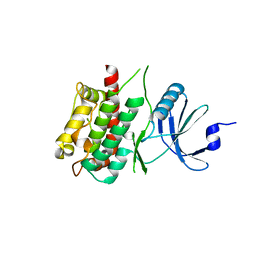 | |
2PPY
 
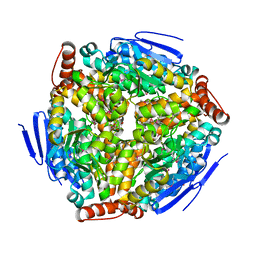 | | Crystal structure of Enoyl-CoA hydrates (gk_1992) from Geobacillus Kaustophilus HTA426 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Kavyashree, M, Sekar, K, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-01 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Enoyl-CoA hydrates (gk_1992) from Geobacillus Kaustophilus HTA426
To be Published
|
|
7QTZ
 
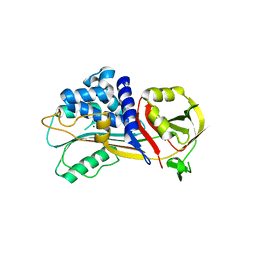 | | Crystal structure of Iripin-1 serpin from tick Ixodes ricinus | | Descriptor: | MAGNESIUM ION, Putative salivary serpin | | Authors: | Kascakova, B, Kuta Smatanova, I, Chmelar, J, Prudnikova, T. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iripin-1, a new anti-inflammatory tick serpin, inhibits leukocyte recruitment in vivo while altering the levels of chemokines and adhesion molecules.
Front Immunol, 14, 2023
|
|
3KPY
 
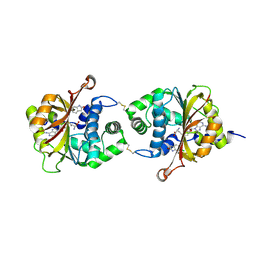 | | Crystal Structure of hPNMT in Complex AdoHcy and 6-Chlorooxindole | | Descriptor: | 6-chloro-1,3-dihydro-2H-indol-2-one, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
6NNF
 
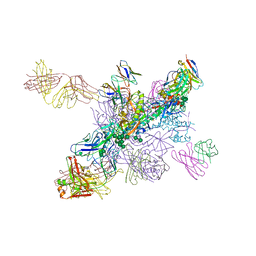 | |
6DOS
 
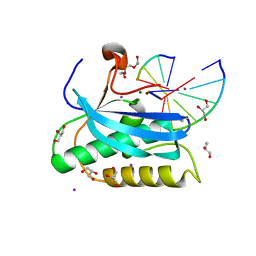 | |
6DMV
 
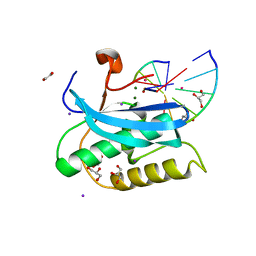 | |
6DPH
 
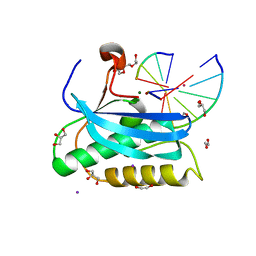 | |
8JST
 
 | |
5ZRL
 
 | | M. smegmatis antimutator protein MutT2 in complex with CDP | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-DIPHOSPHATE, Putative mutator protein MutT2/NUDIX hydrolase | | Authors: | Singh, A, Arif, S.M, Sang, P.B, Varshney, U, Vijayan, M. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the specificity and catalytic mechanism of mycobacterial nucleotide pool sanitizing enzyme MutT2.
J.Struct.Biol., 204, 2018
|
|
6NF2
 
 | |
5EXI
 
 | | Crystal structure of M. tuberculosis lipoyl synthase at 2.28 A resolution | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, IRON/SULFUR CLUSTER, Lipoyl synthase | | Authors: | McLaughlin, M.I, Lanz, N.D, Goldman, P.J, Lee, K.-H, Booker, S.J, Drennan, C.L. | | Deposit date: | 2015-11-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystallographic snapshots of sulfur insertion by lipoyl synthase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6TDH
 
 | | Crystal structure of Aspergillus fumigatus Glucosamine-6-phosphate N-acetyltransferase 1 in complex with compound 1 | | Descriptor: | 3-(2-chlorophenyl)-4~{H}-1,2,4-triazole, ACETYL COENZYME *A, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Raimi, O.G, Stanley, M, Lockhart, D. | | Deposit date: | 2019-11-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting a critical step in fungal hexosamine biosynthesis.
J.Biol.Chem., 295, 2020
|
|
6DOQ
 
 | |
