1QGZ
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LEU 78 REPLACED BY ASP (L78D) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1999-05-10 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of a cluster of hydrophobic residues near the FAD cofactor in Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal complex formation and electron transfer to ferredoxin.
J.Biol.Chem., 276, 2001
|
|
1QIE
 
 | |
1QIS
 
 | | ASPARTATE AMINOTRANSFERASE FROM ESCHERICHIA COLI, C191F MUTATION, WITH BOUND MALEATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, MALEIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1999-06-15 | | Release date: | 2000-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Role of Residues Outside the Active Site in Catalysis: Structural Basis for Function of C191 Mutants of E. Coli Aspartate Aminotransferase
Protein Eng., 13, 2000
|
|
1QAM
 
 | | THE STRUCTURE OF THE RRNA METHYLTRANSFERASE ERMC': IMPLICATIONS FOR THE REACTION MECHANISM | | Descriptor: | ACETATE ION, ERMC' METHYLTRANSFERASE | | Authors: | Schluckebier, G, Zhong, P, Stewart, K.D, Kavanaugh, T.J, Abad-Zapatero, C. | | Deposit date: | 1999-03-25 | | Release date: | 2000-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A structure of the rRNA methyltransferase ErmC' and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism.
J.Mol.Biol., 289, 1999
|
|
1QAV
 
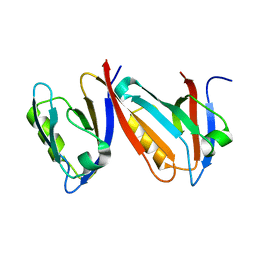 | | Unexpected Modes of PDZ Domain Scaffolding Revealed by Structure of NNOS-Syntrophin Complex | | Descriptor: | ALPHA-1 SYNTROPHIN (RESIDUES 77-171), NEURONAL NITRIC OXIDE SYNTHASE (RESIDUES 1-130) | | Authors: | Hillier, B.J, Christopherson, K.S, Prehoda, K.E, Bredt, D.S, Lim, W.A. | | Deposit date: | 1999-03-30 | | Release date: | 1999-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex.
Science, 284, 1999
|
|
1H5Z
 
 | |
1H3Y
 
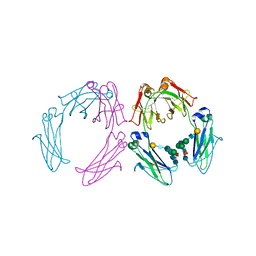 | | Crystal structure of a human IgG1 Fc-fragment,high salt condition | | Descriptor: | IG GAMMA-1 CHAIN C REGION, alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Krapp, S, Mimura, Y, Jefferis, R, Huber, R, Sondermann, P. | | Deposit date: | 2002-09-19 | | Release date: | 2003-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Analysis of Human Igg-Fc Glycoforms Reveals a Correlation between Glycosylation and Structural Integrity.
J.Mol.Biol., 325, 2003
|
|
1HH8
 
 | | The active N-terminal region of p67phox: Structure at 1.8 Angstrom resolution and biochemical characterizations of the A128V mutant implicated in chronic granulomatous disease | | Descriptor: | CITRATE ANION, NEUTROPHIL CYTOSOL FACTOR 2 | | Authors: | Grizot, S, Fieschi, F, Dagher, M.-C, Pebay-Peyroula, E. | | Deposit date: | 2000-12-21 | | Release date: | 2001-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Active N-Terminal Region of P67Phox: Structure at 1.8 Angstrom Resolution and Biochemical Characterizations of the A128V Mutant Implicated in Chronic Granulomatous Disease
J.Biol.Chem., 276, 2001
|
|
1GZ3
 
 | | Molecular mechanism for the regulation of human mitochondrial NAD(P)+-dependent malic enzyme by ATP and fumarate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FUMARIC ACID, MANGANESE (II) ION, ... | | Authors: | Yang, Z, Lanks, C.W, Tong, L. | | Deposit date: | 2002-05-14 | | Release date: | 2003-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Mechanism for the Regulation of Human Mitochondrial Nad(P)+-Dependent Malic Enzyme by ATP and Fumarate
Structure, 10, 2002
|
|
1HE9
 
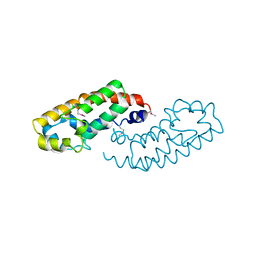 | | Crystal structure of the GAP domain of the Pseudomonas aeruginosa ExoS toxin | | Descriptor: | EXOENZYME S | | Authors: | Wurtele, M, Renault, L, Barbieri, J.T, Wittinghofer, A, Wolf, E. | | Deposit date: | 2000-11-21 | | Release date: | 2001-03-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Exos Gtpase Activating Domain
FEBS Lett., 491, 2001
|
|
4BF4
 
 | |
4BJ8
 
 | | Zebavidin | | Descriptor: | BIOTIN, GLYCEROL, ZEBAVIDIN | | Authors: | Airenne, T.T, Parthiban, M, Niederhauser, B, Zmurko, J, Kulomaa, M.S, Hytonen, V.P, Johnson, M.S. | | Deposit date: | 2013-04-17 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Zebavidin
Plos One, 8, 2013
|
|
3GYR
 
 | | Structure of Phenoxazinone synthase from Streptomyces antibioticus reveals a new type 2 copper center. | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, GLYCEROL, ... | | Authors: | Smith, A.W, Camara-Artigas, A, Wang, M, Francisco, W.A, Allen, J.P. | | Deposit date: | 2009-04-05 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Phenoxazinone Synthase from Streptomyces Antibioticus Reveals a New Type 2 Copper Center.
Biochemistry, 45, 2006
|
|
3GQQ
 
 | | Crystal structure of the human retinal protein 4 (unc-119 homolog A). Northeast Structural Genomics Consortium target HR3066a | | Descriptor: | Protein unc-119 homolog A, UNKNOWN LIGAND | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Shastry, R, Foote, E.L, Ciccosanti, C, Sahdev, S, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-24 | | Release date: | 2009-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Crystal structure of the human retinal protein 4 (unc-119 homolog A).
To be Published
|
|
4C2M
 
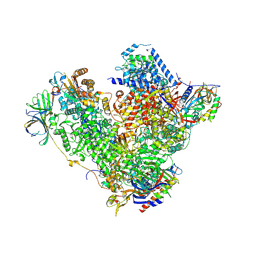 | | Structure of RNA polymerase I at 2.8 A resolution | | Descriptor: | DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA12, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA135, DNA-DIRECTED RNA POLYMERASE I SUBUNIT RPA14, ... | | Authors: | Engel, C, Sainsbury, S, Cheung, A.C, Kostrewa, D, Cramer, P. | | Deposit date: | 2013-08-19 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Polymerase I Structure and Transcription Regulation.
Nature, 502, 2013
|
|
4BNJ
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-methyl- 2-phenoxyphenol | | Descriptor: | 5-methyl-2-phenoxyphenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
3GXI
 
 | | Crystal structure of acid-beta-glucosidase at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, PHOSPHATE ION | | Authors: | Lieberman, R.L. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Effects of pH and iminosugar pharmacological chaperones on lysosomal glycosidase structure and stability.
Biochemistry, 48, 2009
|
|
4BZ7
 
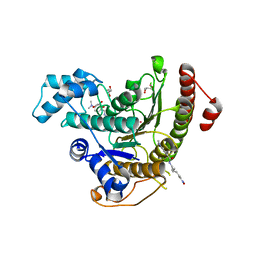 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, GLYCEROL, HISTONE DEACETYLASE 8, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
3HDQ
 
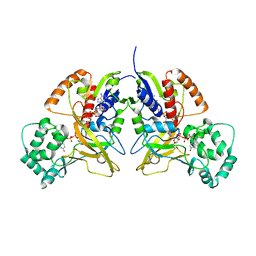 | |
3HE3
 
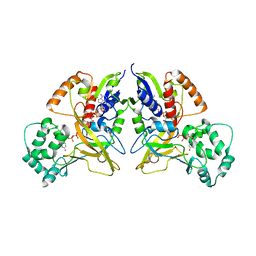 | |
3HDY
 
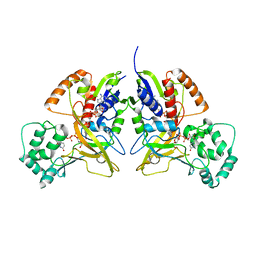 | | Crystal Structure of UDP-galactopyranose mutase (reduced form) in complex with substrate | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Partha, S.K, van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of substrate binding to UDP-galactopyranose mutase: crystal structures in the reduced and oxidized state complexed with UDP-galactopyranose and UDP.
J.Mol.Biol., 394, 2009
|
|
3HEA
 
 | | The L29P/L124I mutation of Pseudomonas fluorescens esterase | | Descriptor: | Arylesterase, ETHYL ACETATE, GLYCEROL, ... | | Authors: | Kazlauskas, R.J, Schrag, J.D, Cheeseman, J.D, Morley, K.L. | | Deposit date: | 2009-05-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Switching catalysis from hydrolysis to perhydrolysis in Pseudomonas fluorescens esterase.
Biochemistry, 49, 2010
|
|
4ALK
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 5-ethyl- 2-phenoxyphenol | | Descriptor: | 5-ETHYL-2-PHENOXYPHENOL, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Tonge, P.J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
3IGI
 
 | | Tertiary Architecture of the Oceanobacillus Iheyensis Group II Intron | | Descriptor: | 5'-R(*CP*GP*CP*UP*CP*UP*AP*CP*UP*CP*UP*AP*U)-3', Group IIC intron, MAGNESIUM ION, ... | | Authors: | Toor, N, Keating, K.S, Fedorova, O, Rajashankar, K, Wang, J, Pyle, A.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | Tertiary architecture of the Oceanobacillus iheyensis group II intron.
Rna, 16, 2010
|
|
3FXI
 
 | | Crystal structure of the human TLR4-human MD-2-E.coli LPS Ra complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-HYDROXY-TETRADECANOIC ACID, ... | | Authors: | Park, B.S, Song, D.H, Kim, H.M, Lee, J.-O. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex
Nature, 458, 2009
|
|
