1DGZ
 
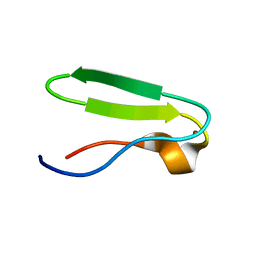 | | RIBOSMAL PROTEIN L36 FROM THERMUS THERMOPHILUS: NMR STRUCTURE ENSEMBLE | | Descriptor: | PROTEIN (L36 RIBOSOMAL PROTEIN), ZINC ION | | Authors: | Hard, T, Rak, A, Allard, P, Kloo, L, Garber, M. | | Deposit date: | 1999-11-27 | | Release date: | 1999-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ribosomal protein L36 from Thermus thermophilus reveals a zinc-ribbon-like fold.
J.Mol.Biol., 296, 2000
|
|
2Y7H
 
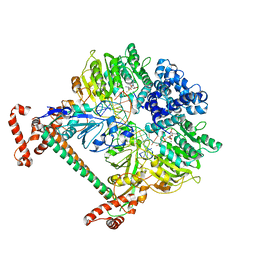 | | Atomic model of the DNA-bound methylase complex from the Type I restriction-modification enzyme EcoKI (M2S1). Based on fitting into EM map 1534. | | Descriptor: | 5'-D(*GP*TP*TP*CP*AP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*GP*CP*AP*AP*C)-3', 5'-D(*GP*TP*TP*GP*CP*AP*CP*GP*TP*CP*GP*AP*CP*GP *TP*TP*GP*AP*AP*C)-3', S-ADENOSYLMETHIONINE, ... | | Authors: | Kennaway, C.K, Obarska-Kosinska, A, White, J.H, Tuszynska, I, Cooper, L.P, Bujnicki, J.M, Trinick, J, Dryden, D.T.F. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | The Structure of M.Ecoki Type I DNA Methyltransferase with a DNA Mimic Antirestriction Protein.
Nucleic Acids Res., 37, 2009
|
|
2YHC
 
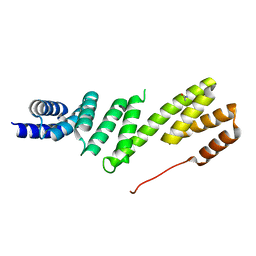 | | Structure of BamD from E. coli | | Descriptor: | UPF0169 LIPOPROTEIN YFIO, UREA | | Authors: | Zeth, K, Albrecht, R. | | Deposit date: | 2011-04-28 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Outer Membrane Protein Biogenesis in Bacteria.
J.Biol.Chem., 286, 2011
|
|
2YH9
 
 | |
1E5V
 
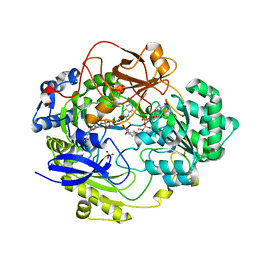 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-03 | | Release date: | 2000-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
2YUE
 
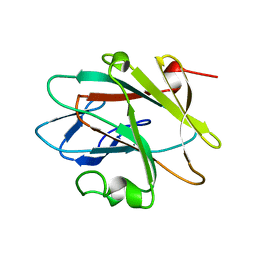 | | Solution structure of the NEUZ (NHR) domain in Neuralized from Drosophila melanogaster | | Descriptor: | Protein neuralized | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of the NHR1 domain of the Drosophila neuralized E3 ligase in the notch signaling pathway.
J.Mol.Biol., 393, 2009
|
|
2BGK
 
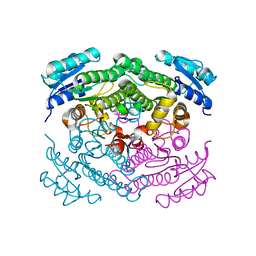 | | X-Ray structure of apo-Secoisolariciresinol Dehydrogenase | | Descriptor: | RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
2YSZ
 
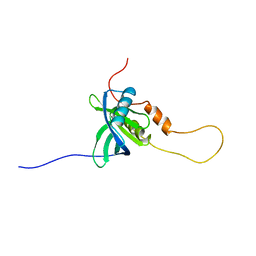 | | Solution structure of the chimera of the C-terminal PID domain of Fe65L and the C-terminal tail peptide of APP | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 2 and Amyloid beta A4 protein | | Authors: | Li, H, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal phosphotyrosine interaction domain of Fe65L1 complexed with the cytoplasmic tail of amyloid precursor protein reveals a novel peptide binding mode
J.Biol.Chem., 283, 2008
|
|
2HS3
 
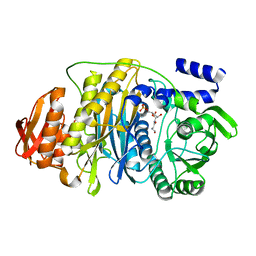 | | T. maritima PurL complexed with FGAR | | Descriptor: | N-(N-FORMYLGLYCYL)-5-O-PHOSPHONO-BETA-D-RIBOFURANOSYLAMINE, PHOSPHATE ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
1EOV
 
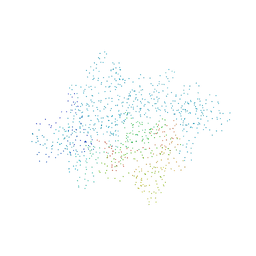 | | FREE ASPARTYL-TRNA SYNTHETASE (ASPRS) (E.C. 6.1.1.12) FROM YEAST | | Descriptor: | ASPARTYL-TRNA SYNTHETASE | | Authors: | Sauter, C, Lorber, B, Cavarelli, J, Moras, D, Giege, R. | | Deposit date: | 2000-03-24 | | Release date: | 2000-09-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free yeast aspartyl-tRNA synthetase differs from the tRNA(Asp)-complexed enzyme by structural changes in the catalytic site, hinge region, and anticodon-binding domain.
J.Mol.Biol., 299, 2000
|
|
4NVG
 
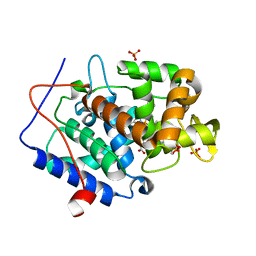 | |
4NVJ
 
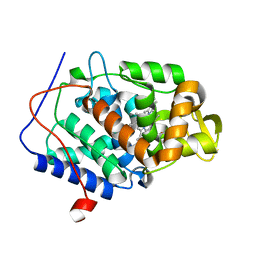 | |
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2HRU
 
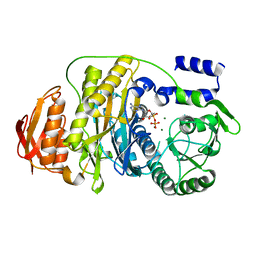 | | T. maritima PurL complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase II | | Authors: | Ealick, S.E, Morar, M. | | Deposit date: | 2006-07-20 | | Release date: | 2007-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complexed Structures of Formylglycinamide Ribonucleotide Amidotransferase from Thermotoga maritima Describe a Novel ATP Binding Protein Superfamily
Biochemistry, 45, 2006
|
|
8R3G
 
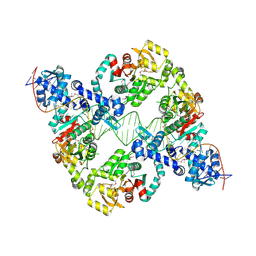 | | Central glycolytic genes regulator (CggR) bound to DNA operator | | Descriptor: | Central glycolytic genes regulator, operator DNA | | Authors: | Skerlova, J, Soltysova, M, Rezacova, P, Skubnik, K. | | Deposit date: | 2023-11-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural characterization of two prototypical repressors of SorC family reveals tetrameric assemblies on DNA and mechanism of function.
Nucleic Acids Res., 52, 2024
|
|
4O0U
 
 | |
2C6E
 
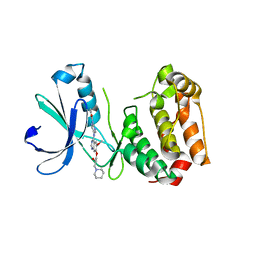 | | Aurora A kinase activated mutant (T287D) in complex with a 5- aminopyrimidinyl quinazoline inhibitor | | Descriptor: | N-{5-[(7-{[(2S)-2-HYDROXY-3-PIPERIDIN-1-YLPROPYL]OXY}-6-METHOXYQUINAZOLIN-4-YL)AMINO]PYRIMIDIN-2-YL}BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Pauptit, R.A, Pannifer, A.D, Breed, J, McMiken, H.H.J, Rowsell, S, Anderson, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-01-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sar and Inhibitor Complex Structure Determination of a Novel Class of Potent and Specific Aurora Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2BK2
 
 | | The prepore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
1EV0
 
 | |
4NC4
 
 | | Crystal structure of photoreceptor AtUVR8 mutant W285F and light-induced structural changes at 120K | | Descriptor: | MAGNESIUM ION, Ultraviolet-B receptor UVR8 | | Authors: | Yang, X, Zeng, X, Zhao, K.-H, Ren, Z. | | Deposit date: | 2013-10-23 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamic Crystallography Reveals Early Signalling Events in Ultraviolet Photoreceptor UVR8.
Nat Plants, 1, 2015
|
|
1EGX
 
 | | SOLUTION STRUCTURE OF THE ENA-VASP HOMOLOGY 1 (EVH1) DOMAIN OF HUMAN VASODILATOR-STIMULATED PHOSPHOPROTEIN (VASP) | | Descriptor: | VASODILATOR-STIMULATED PHOSPHOPROTEIN | | Authors: | Ball, L, Kuhne, R, Hoffmann, B, Hafner, A, Schmieder, P, Volkmer-Engert, R, Hof, M, Wahl, M, Schneider-Mergener, J, Walter, U, Oschkinat, H, Jarchau, T. | | Deposit date: | 2000-02-17 | | Release date: | 2000-09-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity.
EMBO J., 19, 2000
|
|
2BGM
 
 | | X-Ray structure of ternary-Secoisolariciresinol Dehydrogenase | | Descriptor: | MATAIRESINOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), RHIZOME SECOISOLARICIRESINOL DEHYDROGENASE | | Authors: | Youn, B, Moinuddin, S.G, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2004-12-23 | | Release date: | 2005-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Apo-Form and Binary/Ternary Complexes of Podophyllum Secoisolariciresinol Dehydrogenase, an Enzyme Involved in Formation of Health-Protecting and Plant Defense Lignans
J.Biol.Chem., 280, 2005
|
|
1EGP
 
 | |
2BUG
 
 | | Solution structure of the TPR domain from Protein phosphatase 5 in complex with Hsp90 derived peptide | | Descriptor: | HSP90, SERINE/THREONINE PROTEIN PHOSPHATASE 5 | | Authors: | Cliff, M.J, Harris, R, Barford, D, Ladbury, J.E, Williams, M.A. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-16 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | Conformational Diversity in the Tpr Domain-Mediated Interaction of Protein Phosphatase 5 with Hsp90.
Structure, 14, 2006
|
|
4NVF
 
 | |
