7ZNS
 
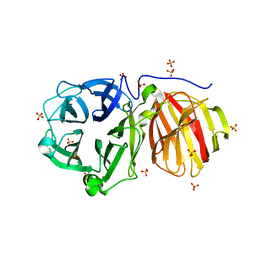 | |
6JCH
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Orthorhombic form | | Descriptor: | Pilus assembly protein, SODIUM ION | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-01-28 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.536 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
6D9M
 
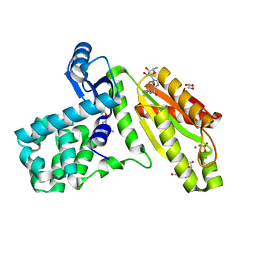 | | T4-Lysozyme fusion to Geobacter GGDEF | | Descriptor: | ACETATE ION, Fusion protein of Endolysin,Response receiver sensor diguanylate cyclase, GAF domain-containing, ... | | Authors: | Hallberg, Z, Doxzen, K, Kranzusch, P.J, Hammond, M. | | Deposit date: | 2018-04-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and mechanism of a Hypr GGDEF enzyme that activates cGAMP signaling to control extracellular metal respiration.
Elife, 8, 2019
|
|
7AQ9
 
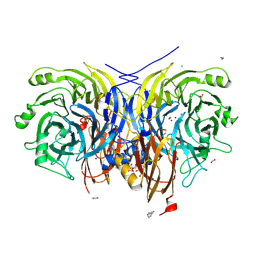 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583W | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.585 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7RAH
 
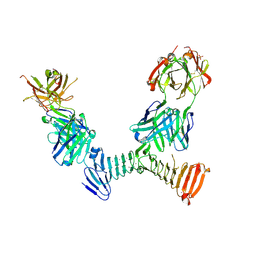 | | Adenylate cyclase toxin RTX domain fragment bound to M1H5 Fab and M2B10 Fab | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA,Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, M1H5 Fab Heavy Chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2021-07-01 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for antibody binding to adenylate cyclase toxin reveals RTX linkers as neutralization-sensitive epitopes.
Plos Pathog., 17, 2021
|
|
6DA1
 
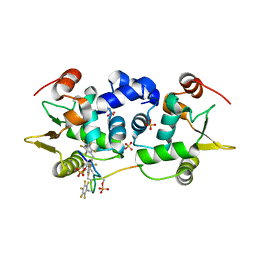 | | ETS1 in complex with synthetic SRR mimic | | Descriptor: | Protein C-ets-1, SULFATE ION, serine-rich region (SRR) peptide | | Authors: | Perez-Borrajero, C, Okon, M, Lin, C.S, Scheu, K, Murphy, M.E.P, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.000127 Å) | | Cite: | The Biophysical Basis for Phosphorylation-Enhanced DNA-Binding Autoinhibition of the ETS1 Transcription Factor.
J. Mol. Biol., 431, 2019
|
|
8A45
 
 | |
8A4D
 
 | |
6D48
 
 | | Cell Surface Receptor | | Descriptor: | Myeloid cell surface antigen CD33 | | Authors: | Hermans, S.J, Miles, L.A, Parker, M.W. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Small Molecule Binding to Alzheimer Risk Factor CD33 Promotes A beta Phagocytosis.
Iscience, 19, 2019
|
|
7B34
 
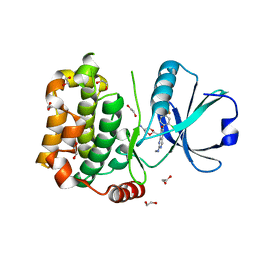 | | MST3 in complex with compound MRIA12 | | Descriptor: | 1,2-ETHANEDIOL, 8-[(5-azanyl-1,3-dioxan-2-yl)methyl]-6-[2-chloranyl-4-(3-fluoranyl-6-methyl-pyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
6D41
 
 | | Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D4V
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 22 (VCC061422) | | Descriptor: | 2-cyclohexyl-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
8A33
 
 | |
8T2B
 
 | |
7AX2
 
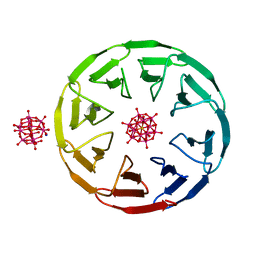 | | Crystal structure of the computationally designed Scone-E protein co-crystallized with STA, form b | | Descriptor: | Keggin (STA), Monolacunary Keggin (STA), SODIUM ION, ... | | Authors: | Mylemans, B, Vandebroek, L, Parac-Vogt, T.N, Voet, A.R.D. | | Deposit date: | 2020-11-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Scone: pseudosymmetric folding of a symmetric designer protein.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7RMR
 
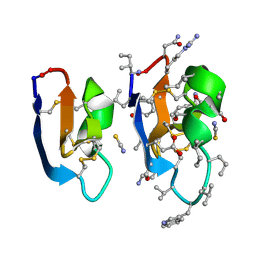 | | Crystal structure of [I11L]cycloviolacin O2 | | Descriptor: | D-[I11L]cycloviolacin O2, THIOCYANATE ION, [I11L]cycloviolacin O2 | | Authors: | Huang, Y.H, Du, Q, Craik, D.J. | | Deposit date: | 2021-07-28 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Enabling Efficient Folding and High-Resolution Crystallographic Analysis of Bracelet Cyclotides.
Molecules, 26, 2021
|
|
8T2O
 
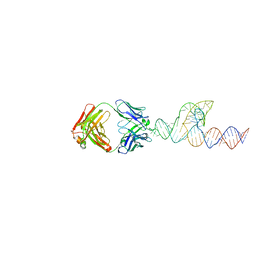 | |
8T2A
 
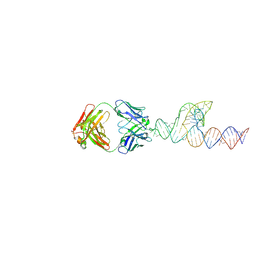 | |
8A5K
 
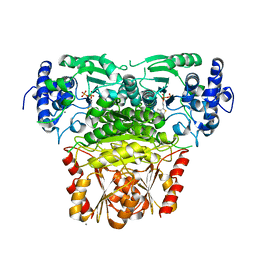 | |
7RIJ
 
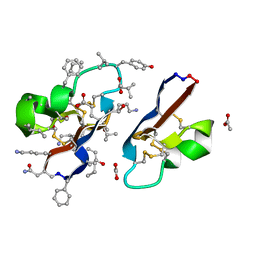 | | [I11G]hyen D | | Descriptor: | ACETATE ION, Cyclotide hyen-D, D-[I11L]hyen D | | Authors: | Du, Q, Huang, Y.H, Wang, C.K, Craik, D.J. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Enabling efficient folding and high-resolution crystallographic analysis of bracelet cyclotides
Molecules, 26(18), 2021
|
|
7AF1
 
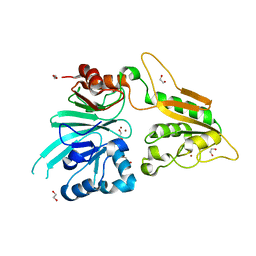 | | The structure of Artemis/SNM1C/DCLRE1C with 2 Zinc ions | | Descriptor: | 1,2-ETHANEDIOL, Protein artemis, ZINC ION | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-19 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
7RIH
 
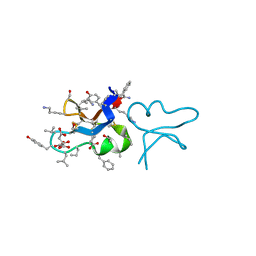 | | hyen D | | Descriptor: | CITRATE ANION, Cyclotide hyen-D, D-[I11L]hyen D | | Authors: | Du, Q, Huang, Y.H, Craik, D.J, Wang, C.K. | | Deposit date: | 2021-07-20 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enabling efficient folding and high-resolution crystallographic analysis of bracelet cyclotides
Molecules, 26(18), 2021
|
|
8A37
 
 | |
7B1I
 
 | |
8T29
 
 | | Crystal structure of SCV PTE RNA in complex with Fab BL3-6 | | Descriptor: | BL3-6 Fab heavy chain, BL3-6 Fab light chain, RNA (90-MER) | | Authors: | Ojha, M, Koirala, D. | | Deposit date: | 2023-06-05 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structure of saguaro cactus virus 3' translational enhancer mimics 5' cap for eIF4E binding.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
