2LU0
 
 | | NMR solution structure of the kappa-zeta region of S.cerevisiae group II intron ai5(gamma) | | Descriptor: | RNA (49-MER) | | Authors: | Donghi, D, Pechlaner, M, Finazzo, C, Knobloch, B, Sigel, R.K.O. | | Deposit date: | 2012-06-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural stabilization of the kappa three-way junction by Mg(II) represents the first step in the folding of a group II intron.
Nucleic Acids Res., 41, 2013
|
|
2LWJ
 
 | |
5ZUB
 
 | |
2M58
 
 | | Structure of 2'-5' AG1 lariat forming ribozyme in its inactive state | | Descriptor: | RNA (59-MER) | | Authors: | Carlomagno, T, Amata, I, Codutti, L, Falb, M, Fohrer, J, Simon, B. | | Deposit date: | 2013-02-18 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural principles of RNA catalysis in a 2'-5' lariat-forming ribozyme.
J.Am.Chem.Soc., 135, 2013
|
|
2M1L
 
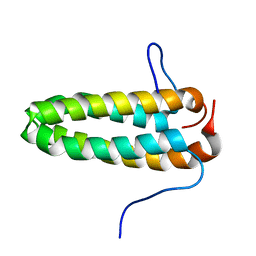 | | Solution NMR Structure of Cyclin-dependent kinase 2-associated protein 2 (CDK2AP2, DOC-1R) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8910C | | Descriptor: | Cyclin-dependent kinase 2-associated protein 2 | | Authors: | Ertekin, A, Janjua, H, Kohan, E, Shastry, R, Pederson, K, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CDK2-associated protein 2 (CDK2AP2, Deleted in Oral Cancer 1 Related protein, DOC-1R)
To be Published
|
|
2M6V
 
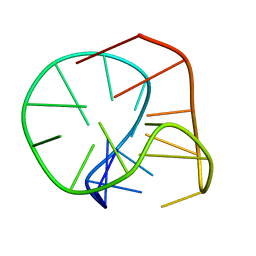 | |
2M3M
 
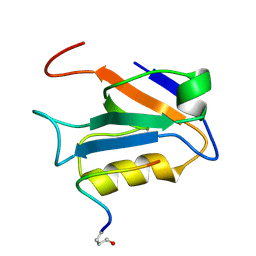 | |
5ZZ8
 
 | | Structure of the Herpes simplex virus type 2 C-capsid with capsid-vertex-specific component | | Descriptor: | Major capsid protein, UL17, UL25, ... | | Authors: | Wang, J.L, Yuan, S, Zhu, D.J, Tang, H, Wang, N, Chen, W.Y, Gao, Q, Li, Y.H, Wang, J.Z, Liu, H.R, Zhang, X.Z, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component.
Nat Commun, 9, 2018
|
|
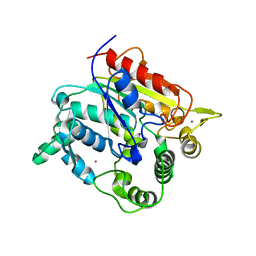
6A07
 
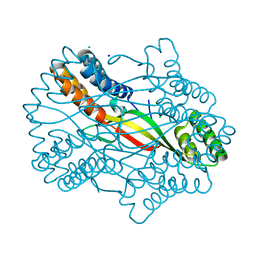 | |
6A12
 
 | |
4XI9
 
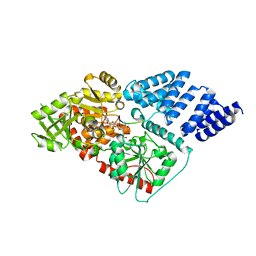 | | Human OGT in complex with UDP-5S-GlcNAc and substrate peptide (RBL2) | | Descriptor: | (2S,3R,4R,5S,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, Retinoblastoma-like protein 2, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Schimpl, M, van Aalten, D.M.F. | | Deposit date: | 2015-01-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The active site of O-GlcNAc transferase imposes constraints on substrate sequence.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XBM
 
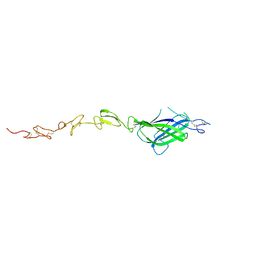 | | X-ray crystal structure of Notch ligand Delta-like 1 | | Descriptor: | Delta-like protein 1, alpha-L-fucopyranose | | Authors: | Kershaw, N.J, Burgess, A.W, Church, N.L, Luo, C.S, Adam, T.E. | | Deposit date: | 2014-12-17 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Notch ligand delta-like1: X-ray crystal structure and binding affinity.
Biochem.J., 468, 2015
|
|
6AAO
 
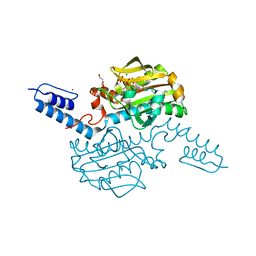 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with TCO*Lys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.403 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
6AB0
 
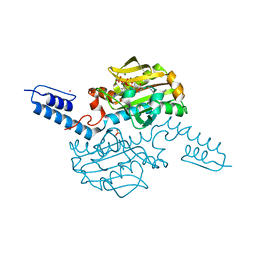 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with pAmPyLys | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N6-{[(6-aminopyridin-3-yl)methoxy]carbonyl}-L-lysine, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-19 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.441 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
5ZRE
 
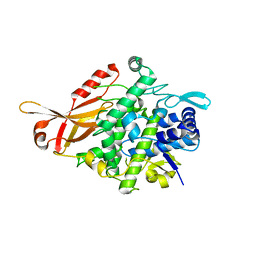 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at high pH condition | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN ATOM, ... | | Authors: | Lee, S, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
2LV2
 
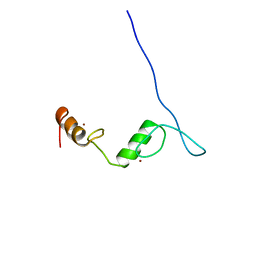 | | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural Genomics Consortium Target HR7614B | | Descriptor: | Insulinoma-associated protein 1, ZINC ION | | Authors: | Yang, Y, Ramelot, T.A, Cort, J.R, Shastry, R, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of C2H2-type Zinc-fingers 4 and 5 from human Insulinoma-associated protein 1 (fragment 424-497), Northeast Structural
Genomics Consortium Target HR7614B (CASP Target)
To be Published
|
|
2LV0
 
 | |
2LW1
 
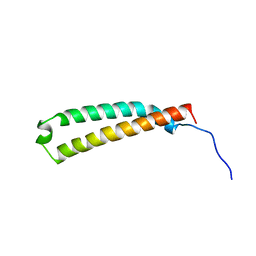 | | The C-terminal domain of the Uup protein is a DNA-binding coiled coil motif | | Descriptor: | ABC transporter ATP-binding protein uup | | Authors: | Carlier, L, Haase, A.S, Burgos Zepeda, M.Y, Dassa, E, Lequin, O. | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of the Uup protein is a DNA-binding coiled coil motif.
J.Struct.Biol., 180, 2012
|
|
2M3V
 
 | |
6A9D
 
 | |
4XJX
 
 | | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HsdR, MAGNESIUM ION | | Authors: | Baikova, T, Stsiapanava, A, Moche, M, Degtjarik, O, Kuta-Smatanova, I, Ettrich, R. | | Deposit date: | 2015-01-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | STRUCTURE OF MUTANT (E165H) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP
To Be Published
|
|
6AA2
 
 | | X-ray structure of ReQy1 (oxidized form) | | Descriptor: | Green fluorescent protein | | Authors: | Sugiura, K, Yasuda, A, Tabushi, N, Tanaka, H, Kurisu, G, Hisabori, T. | | Deposit date: | 2018-07-17 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multicolor redox sensor proteins can visualize redox changes in various compartments of the living cell.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
4Y2D
 
 | | Crystal structure of the mCD1d/7DW8-5/iNKTCR ternary complex | | Descriptor: | 11-(4-fluorophenyl)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]undecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Yu, E.D. | | Deposit date: | 2015-02-09 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural modifications of alphaGalCer in both lipid and carbohydrate moiety influence activation of murine and human iNKT cells
To Be Published
|
|
6AAP
 
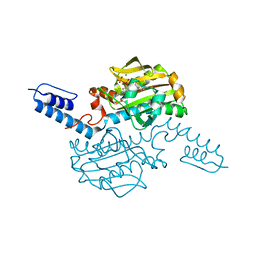 | | Crystal structure of Methanosarcina mazei PylRS(Y306A/Y384F) complexed with ZaeSeCys | | Descriptor: | 3-[(2-{[(benzyloxy)carbonyl]amino}ethyl)selanyl]-L-alanine, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yanagisawa, T, Kuratani, M, Yokoyama, S. | | Deposit date: | 2018-07-18 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell Chem Biol, 26, 2019
|
|
4XNF
 
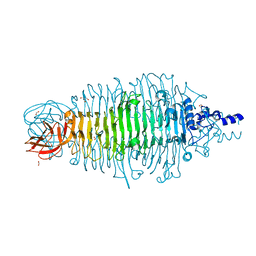 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | Authors: | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | Deposit date: | 2015-01-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
