8OST
 
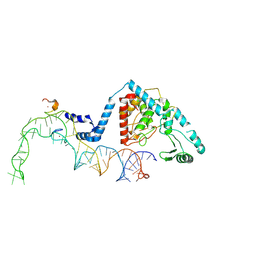 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | Descriptor: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | Authors: | Gilbert, R.J, Yi, G, Ye, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 2024
|
|
8OZN
 
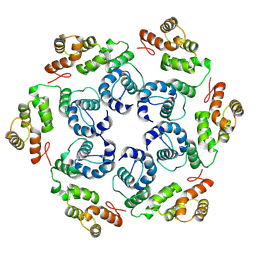 | |
8OZM
 
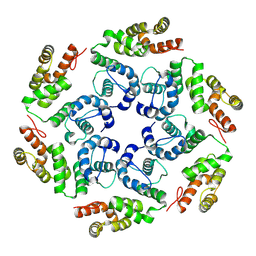 | |
8OJG
 
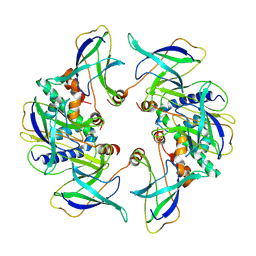 | | Structure of the MlaCD complex (2:6 stoichiometry) | | Descriptor: | ABC transporter substrate-binding protein, ABC-type organic solvent transporter | | Authors: | Wotherspoon, P, Bui, S, Sridhar, P, Bergeron, J.R.C, Knowles, T.J. | | Deposit date: | 2023-03-24 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.38 Å) | | Cite: | The structure of the MlaC-MlaD complex reveals novel insights into periplasmic phospholipid transport processes
To Be Published
|
|
7Q0S
 
 | | Human GYS1-GYG1 complex inhibited-like state bound to glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Glycogen [starch] synthase, muscle, ... | | Authors: | McCorvie, T.J, Shrestha, L, Froese, D.S, Ferreira, I.M, Yue, W.W. | | Deposit date: | 2021-10-16 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular basis for the regulation of human glycogen synthase by phosphorylation and glucose-6-phosphate.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Q13
 
 | | Human GYS1-GYG1 complex activated state bound to glucose-6-phosphate, uridine diphosphate, and glucose | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, Glycogen [starch] synthase, muscle, ... | | Authors: | McCorvie, T.J, Shrestha, L, Froese, D.S, Ferreira, I.M, Yue, W.W. | | Deposit date: | 2021-10-17 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the regulation of human glycogen synthase by phosphorylation and glucose-6-phosphate.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1AWV
 
 | | CYPA COMPLEXED WITH HVGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-05 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
8OZL
 
 | |
8OZK
 
 | |
8PQQ
 
 | |
1B51
 
 | |
8PSV
 
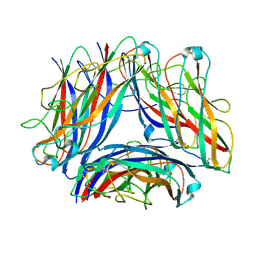 | | 2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M, Zyla, D, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
6OWP
 
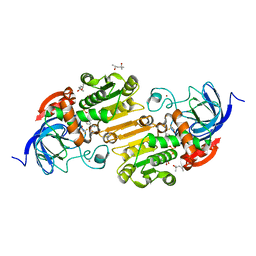 | | Horse liver F93W alcohol dehydrogenase complexed with NAD and trifluoroethanol | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Substitutions of Amino Acid Residues in the Substrate Binding Site of Horse Liver Alcohol Dehydrogenase Have Small Effects on the Structures but Significantly Affect Catalysis of Hydrogen Transfer.
Biochemistry, 59, 2020
|
|
1BHM
 
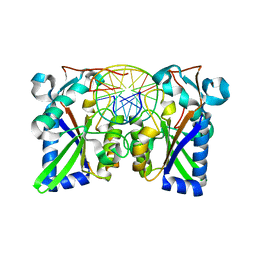 | | RESTRICTION ENDONUCLEASE BAMHI COMPLEX WITH DNA | | Descriptor: | DNA (5'-D(*TP*AP*TP*GP*GP*AP*TP*CP*CP*AP*TP*A)-3'), PROTEIN (BAMHI (E.C.3.1.21.4)) | | Authors: | Aggarwal, A.K, Newman, M. | | Deposit date: | 1995-07-12 | | Release date: | 1995-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding.
Science, 269, 1995
|
|
8PRK
 
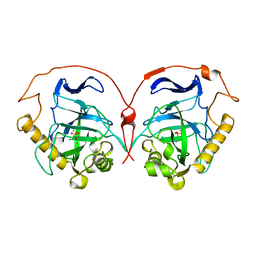 | | THE R78K AND D117E ACTIVE SITE VARIANTS OF SACCHAROMYCES CEREVISIAE SOLUBLE INORGANIC PYROPHOSPHATASE: STRUCTURAL STUDIES AND MECHANISTIC IMPLICATIONS | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, PROTEIN (INORGANIC PYROPHOSPHATASE) | | Authors: | Tuominen, V, Heikinheimo, P, Kajander, T, Torkkel, T, Hyytia, T, Kapyla, J, Lahti, R, Cooperman, B.S, Goldman, A. | | Deposit date: | 1998-09-16 | | Release date: | 1998-12-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The R78K and D117E active-site variants of Saccharomyces cerevisiae soluble inorganic pyrophosphatase: structural studies and mechanistic implications.
J.Mol.Biol., 284, 1998
|
|
8PFC
 
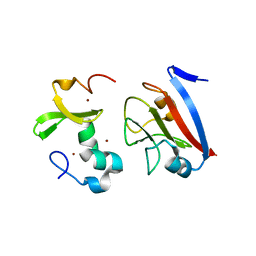 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the zinc finger domain of SPL5 from Arabidopsis thaliana | | Descriptor: | Sequence-variable mosaic (SVM) signal sequence domain-containing protein, Squamosa promoter-binding-like protein 5, ZINC ION | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PKY
 
 | |
8PIF
 
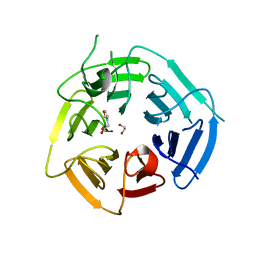 | | Fragment 12 in complex with KLHDC2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(furan-3-yl)ethanoic acid, Kelch domain-containing protein 2 | | Authors: | Boettcher, J, Mayer, M. | | Deposit date: | 2023-06-21 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | KLHDC2 - The Next Level
To Be Published
|
|
8PI2
 
 | |
8PHO
 
 | |
8PM1
 
 | |
8PHT
 
 | |
8PFD
 
 | | Crystal structure of binary complex between Aster yellows witches'-broom phytoplasma effector SAP05 and the von Willebrand Factor Type A domain of the proteasomal ubiquitin receptor Rpn10 from Arabidopsis thaliana | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Huang, W, Liu, Q, Maqbool, A, Stevenson, C.E.M, Lawson, D.M, Kamoun, S, Hogenhout, S.A. | | Deposit date: | 2023-06-15 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Bimodular architecture of bacterial effector SAP05 that drives ubiquitin-independent targeted protein degradation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PFY
 
 | |
8PFV
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with [Ru2Cl(DAniF)(O2CCH3)3] in condition A | | Descriptor: | 9,11-bis(4-methoxyphenyl)-3,7-dimethyl-2,4,6,8-tetraoxa-9,11-diaza-1$l^{4},5$l^{4}-diruthenatricyclo[3.3.3.0^{1,5}]undecane, CHLORIDE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
