1MBV
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ClpSN HETERODIMER TETRAGONAL FORM | | Descriptor: | ATP-Dependent clp Protease ATP-Binding Subunit clp A, Protein yljA | | Authors: | Guo, F, Esser, L, Singh, S.K, Maurizi, M.R, Xia, D. | | Deposit date: | 2002-08-03 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Heterodimeric Complex of the Adaptor, ClpS, with the N-domain of AAA+ Chaperone ClpA
J.Biol.Chem., 277, 2002
|
|
1MC4
 
 | |
1LYL
 
 | |
1MLM
 
 | |
1MMB
 
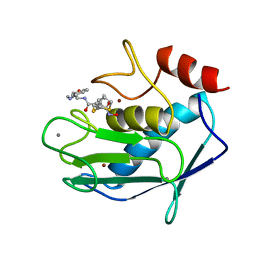 | | COMPLEX OF BB94 WITH THE CATALYTIC DOMAIN OF MATRIX METALLOPROTEINASE-8 | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, MATRIX METALLOPROTEINASE-8, ... | | Authors: | Bode, W, Grams, F. | | Deposit date: | 1995-08-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination and analysis of human neutrophil collagenase complexed with a hydroxamate inhibitor.
Biochemistry, 34, 1995
|
|
1MPF
 
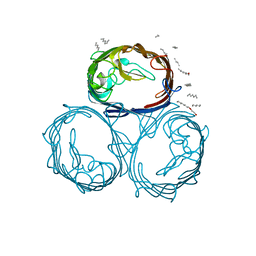 | |
1MPM
 
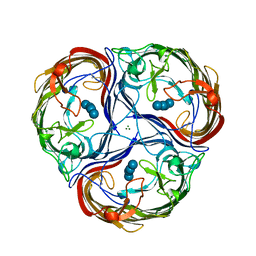 | | MALTOPORIN MALTOSE COMPLEX | | Descriptor: | MAGNESIUM ION, MALTOPORIN, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1996-01-11 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of various maltooligosaccharides bound to maltoporin reveal a specific sugar translocation pathway.
Structure, 4, 1996
|
|
1M8F
 
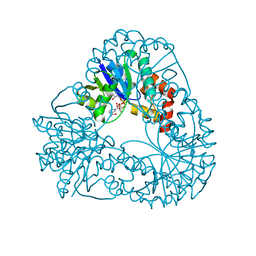 | |
1MA0
 
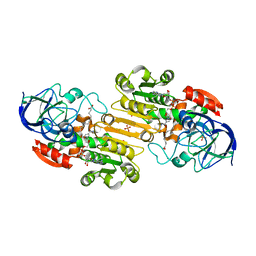 | | Ternary complex of Human glutathione-dependent formaldehyde dehydrogenase with NAD+ and dodecanoic acid | | Descriptor: | Glutathione-dependent formaldehyde dehydrogenase, LAURIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sanghani, P.C, Robinson, H, Bosron, W.F, Hurley, T.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human glutathione-dependent formaldehyde dehydrogenase. Structures of apo, binary, and inhibitory ternary complexes.
Biochemistry, 41, 2002
|
|
1LYS
 
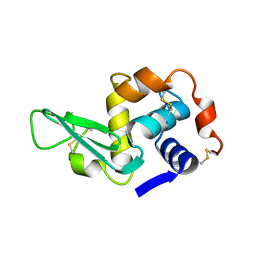 | |
1O7O
 
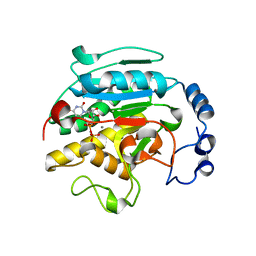 | | Roles of Individual Residues of Alpha-1,3 Galactosyltransferases in Substrate Binding and Catalysis | | Descriptor: | MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, Z, Acharya, K.R, Brew, K. | | Deposit date: | 2002-11-11 | | Release date: | 2003-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Roles of individual enzyme-substrate interactions by alpha-1,3-galactosyltransferase in catalysis and specificity.
Biochemistry, 42, 2003
|
|
1O80
 
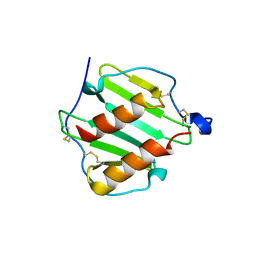 | |
1OA7
 
 | |
1OBB
 
 | | alpha-glucosidase A, AglA, from Thermotoga maritima in complex with maltose and NAD+ | | Descriptor: | ALPHA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lodge, J.A, Maier, T, Liebl, W, Hoffmann, V, Strater, N. | | Deposit date: | 2003-01-29 | | Release date: | 2003-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Thermotoga Maritima Alpha-Glucosidase Agla Defines a New Clan of Nad+-Dependent Glycosidases
J.Biol.Chem., 278, 2003
|
|
1NSN
 
 | |
1NMP
 
 | | Structural genomics, ybgI protein, unknown function | | Descriptor: | Hypothetical protein ybgI, MAGNESIUM ION | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
1NWQ
 
 | | CRYSTAL STRUCTURE OF C/EBPALPHA-DNA COMPLEX | | Descriptor: | 5'-D(*AP*AP*AP*CP*TP*GP*GP*AP*TP*TP*GP*CP*GP*CP*AP*AP*TP*AP*GP*GP*A)-3', 5'-D(*TP*TP*CP*CP*TP*AP*TP*TP*GP*CP*GP*CP*AP*AP*TP*CP*CP*AP*GP*TP*T)-3', CCAAT/enhancer binding protein alpha | | Authors: | Miller, M, Shuman, J.D, Sebastian, T, Dauter, Z, Johnson, P.F. | | Deposit date: | 2003-02-06 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for DNA Recognition by the Basic Region Leucine Zipper
Transcription Factor CCAAT/enhancer Binding Protein Alpha
J.Biol.Chem., 278, 2003
|
|
1NWY
 
 | | COMPLEX OF THE LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS WITH AZITHROMYCIN | | Descriptor: | 23S RIBOSOMAL RRNA, 5S RIBOSOMAL RRNA, AZITHROMYCIN, ... | | Authors: | Schluenzen, F, Harms, J, Franceschi, F, Hansen, H.A.S, Bartels, H, Zarivach, R, Yonath, A. | | Deposit date: | 2003-02-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the antibiotic activity of ketolides and azalides.
Structure, 11, 2003
|
|
1NR0
 
 | | Two Seven-Bladed Beta-Propeller Domains Revealed By The Structure Of A C. elegans Homologue Of Yeast Actin Interacting Protein 1 (AIP1). | | Descriptor: | Actin interacting protein 1, MANGANESE (II) ION | | Authors: | Vorobiev, S.M, Mohri, K, Ono, S, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-23 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of Functional Residues on Caenorhabditis elegans Actin-interacting Protein 1 (UNC-78) for Disassembly of Actin Depolymerizing Factor/Cofilin-bound Actin Filaments
J.Biol.Chem., 279, 2004
|
|
1OAA
 
 | | MOUSE SEPIAPTERIN REDUCTASE COMPLEXED WITH NADP AND OXALOACETATE | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALOACETATE ION, SEPIAPTERIN REDUCTASE, ... | | Authors: | Auerbach, G, Herrmann, A, Bacher, A, Huber, R. | | Deposit date: | 1997-08-25 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
1O3Q
 
 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O45
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU84687. | | Descriptor: | N-ACETYL-N-[1-(1,1'-BIPHENYL-4-YLMETHYL)-2-OXOAZEPAN-3-YL]-3-FORMYL-O-PHOSPHONOTYROSINAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4F
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH RU79073. | | Descriptor: | 1,2,3,4-TETRAHYDROQUINOLIN-8-YL DIHYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O4K
 
 | | CRYSTAL STRUCTURE OF SH2 IN COMPLEX WITH PASBN. | | Descriptor: | BENZYL 2-FORMYLPHENYL HYDROGEN PHOSPHATE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE SRC | | Authors: | Lange, G, Loenze, P, Liesum, A. | | Deposit date: | 2003-06-15 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Requirements for specific binding of low affinity inhibitor fragments to the SH2 domain of (pp60)Src are identical to those for high affinity binding of full length inhibitors.
J.Med.Chem., 46, 2003
|
|
1O7Z
 
 | |
