4J19
 
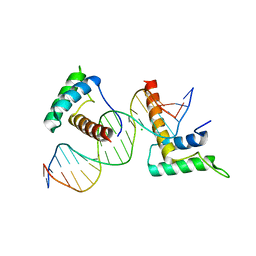 | | Structure of a novel telomere repeat binding protein bound to DNA | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*TP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*CP*A)-3'), ... | | Authors: | Kappei, D, Butter, F, Benda, C, Scheibe, M, Draskovic, I, Stevense, M, Lopes Novo, C, Basquin, C, Krastev, D.B, Kittler, R, Jessberger, R, Londono-Vallejo, A.J, Mann, M, Buchholz, F. | | Deposit date: | 2013-02-01 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment.
Embo J., 32, 2013
|
|
4J0S
 
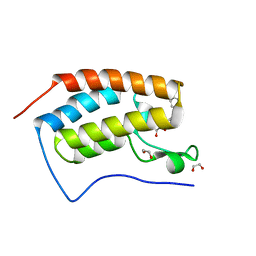 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(S)-hydroxy(phenyl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Hewings, D.S, von Delft, F, Conway, S.J, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Optimization of 3,5-dimethylisoxazole derivatives as potent bromodomain ligands.
J.Med.Chem., 56, 2013
|
|
4MD9
 
 | |
1ZKF
 
 | |
4MHG
 
 | | Crystal structure of ETV6 bound to a specific DNA sequence | | Descriptor: | Complementary Specific 14 bp DNA, Specific 14 bp DNA, Transcription factor ETV6 | | Authors: | Chan, A.C, De, S, Coyne III, H.J, Okon, M, Murphy, M.E, Graves, B.J, McIntosh, L.P. | | Deposit date: | 2013-08-29 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
1ZMZ
 
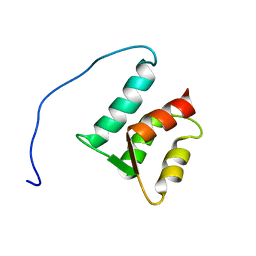 | | Solution structure of the N-terminal domain (M1-S98) of human centrin 2 | | Descriptor: | Centrin-2 | | Authors: | Yang, A, Miron, S, Duchambon, P, Assairi, L, Blouquit, Y, Craescu, C.T. | | Deposit date: | 2005-05-11 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of human centrin 2 has a closed structure, binds calcium with a very low affinity, and plays a role in the protein self-assembly
Biochemistry, 45, 2006
|
|
4KMH
 
 | |
4K1N
 
 | |
1XPA
 
 | | SOLUTION STRUCTURE OF THE DNA-AND RPA-BINDING DOMAIN OF THE HUMAN REPAIR FACTOR XPA, NMR, 1 STRUCTURE | | Descriptor: | XPA, ZINC ION | | Authors: | Ikegami, T, Kuraoka, I, Saijo, M, Kodo, N, Kyogoku, Y, Morikawa, K, Tanaka, K, Shirakawa, M. | | Deposit date: | 1998-07-06 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA- and RPA-binding domain of the human repair factor XPA.
Nat.Struct.Biol., 5, 1998
|
|
4MEN
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 5-methyl-triazolopyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,5-dimethyl-N-(4-methylbenzyl)[1,2,4]triazolo[1,5-a]pyrimidin-7-amine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4MI5
 
 | | Crystal structure of the EZH2 SET domain | | Descriptor: | Histone-lysine N-methyltransferase EZH2, SULFATE ION, ZINC ION | | Authors: | Antonysamy, S, Condon, B, Druzina, Z, Bonanno, J, Gheyi, T, Macewan, I, Zhang, A, Ashok, S, Russell, M, Luz, J.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Context of Disease-Associated Mutations and Putative Mechanism of Autoinhibition Revealed by X-Ray Crystallographic Analysis of the EZH2-SET Domain.
Plos One, 8, 2013
|
|
4KOD
 
 | | Structure of p97 N-D1 R155H mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Xia, D, Tang, W.K. | | Deposit date: | 2013-05-11 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Altered Intersubunit Communication Is the Molecular Basis for Functional Defects of Pathogenic p97 Mutants.
J.Biol.Chem., 288, 2013
|
|
1Z00
 
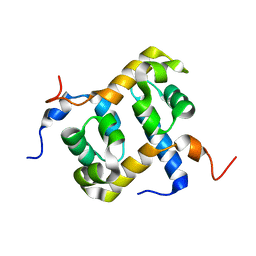 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2005-03-01 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
1XNA
 
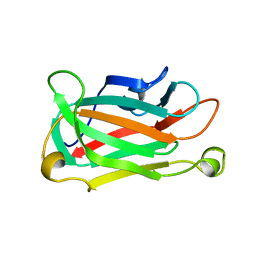 | |
1XNT
 
 | |
4KM9
 
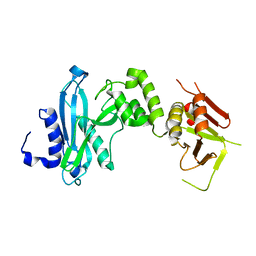 | |
1XJL
 
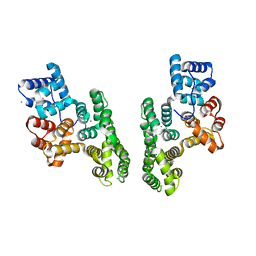 | |
4LY1
 
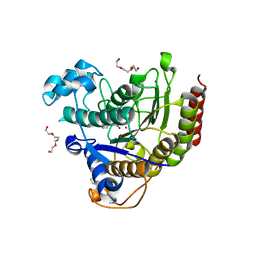 | |
4MEQ
 
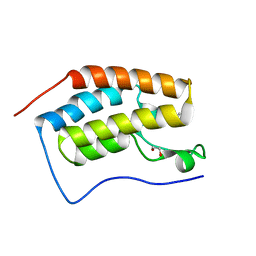 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 5-methyl-triazolopyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-7-phenyl[1,2,4]triazolo[1,5-a]pyrimidin-2-amine, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
1ZAH
 
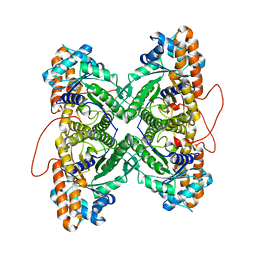 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
1ZNQ
 
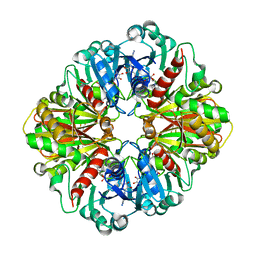 | | Crystal Structure of Human Liver GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, liver, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ismail, S.A, Park, H.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of Human liver GAPDH
To be Published
|
|
4LYS
 
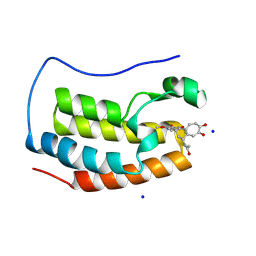 | | Crystal Structure of BRD4(1) bound to Colchiceine | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-10-hydroxy-1,2,3-trimethoxy-9-oxo-5,6,7,9-tetrahydrobenzo[a]heptalen-7-yl]acetamide, SODIUM ION | | Authors: | Wohlwend, D, Gerhardt, S, Einsle, O. | | Deposit date: | 2013-07-31 | | Release date: | 2014-01-15 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 4-Acyl pyrroles: mimicking acetylated lysines in histone code reading.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4MEO
 
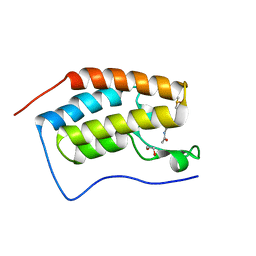 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-methyl-quinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
1YUW
 
 | |
1ZAL
 
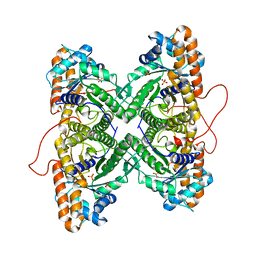 | | Fructose-1,6-bisphosphate aldolase from rabbit muscle in complex with partially disordered tagatose-1,6-bisphosphate, a weak competitive inhibitor | | Descriptor: | Fructose-bisphosphate aldolase A, PHOSPHATE ION | | Authors: | St-Jean, M, Lafrance-Vanasse, J, Liotard, B, Sygusch, J. | | Deposit date: | 2005-04-06 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | High Resolution Reaction Intermediates of Rabbit Muscle Fructose-1,6-bisphosphate Aldolase: substrate cleavage and induced fit.
J.Biol.Chem., 280, 2005
|
|
