3OLV
 
 | |
3OLY
 
 | |
1MB3
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.5 IN COMPLEX WITH MG2+ | | Descriptor: | MAGNESIUM ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1M5U
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK. STRUCTURE AT PH 8.0 IN THE APO-FORM | | Descriptor: | cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus
J.Biol.Chem., 277, 2002
|
|
1M5T
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK | | Descriptor: | cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus
J.Biol.Chem., 277, 2002
|
|
1MB0
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.0 IN COMPLEX WITH MN2+ | | Descriptor: | MANGANESE (II) ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
1MIH
 
 | | A ROLE FOR CHEY GLU 89 IN CHEZ-MEDIATED DEPHOSPHORYLATION OF THE E. COLI CHEMOTAXIS RESPONSE REGULATOR CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, MANGANESE (II) ION, ... | | Authors: | Silversmith, R.E, Guanga, G.P, Betts, L, Chu, C, Zhao, R, Bourret, R.B. | | Deposit date: | 2002-08-23 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89.
J.Bacteriol., 185, 2003
|
|
2LPM
 
 | |
1NAT
 
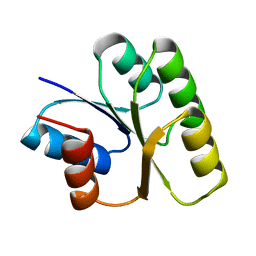 | | CRYSTAL STRUCTURE OF SPOOF FROM BACILLUS SUBTILIS | | Descriptor: | SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Zapf, J, Hoch, J.A, Whiteley, J.M, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1997-09-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A response regulatory protein with the site of phosphorylation blocked by an arginine interaction: crystal structure of Spo0F from Bacillus subtilis.
Biochemistry, 36, 1997
|
|
1NXV
 
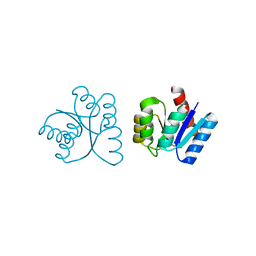 | |
1P6Q
 
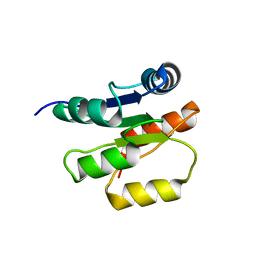 | | NMR Structure of the Response regulator CheY2 from Sinorhizobium meliloti, complexed with Mg++ | | Descriptor: | CheY2 | | Authors: | Riepl, H, Scharf, B, Maurer, T, Schmitt, R, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-04-30 | | Release date: | 2004-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the inactive and BeF3-activated response regulator CheY2.
J.Mol.Biol., 338, 2004
|
|
1PEY
 
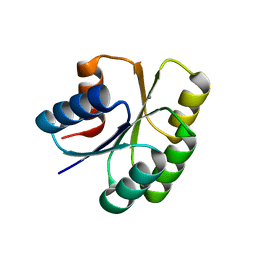 | | Crystal structure of the Response Regulator Spo0F complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, Sporulation initiation phosphotransferase F | | Authors: | Mukhopadhyay, D, Sen, U, Zapf, J, Varughese, K.I. | | Deposit date: | 2003-05-22 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Metals in the sporulation phosphorelay: manganese binding by the response regulator Spo0F.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1YMU
 
 | |
1YMV
 
 | |
1PUX
 
 | | NMR Solution Structure of BeF3-Activated Spo0F, 20 conformers | | Descriptor: | Sporulation initiation phosphotransferase F | | Authors: | Gardino, A.K, Volkman, B.F, Cho, H.S, Lee, S.Y, Wemmer, D.E, Kern, D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of BeF(3)(-)-activated Spo0F reveals the conformational switch in a phosphorelay system.
J.Mol.Biol., 331, 2003
|
|
1NTR
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL RECEIVER DOMAIN OF NTRC | | Descriptor: | NTRC RECEIVER DOMAIN | | Authors: | Volkman, B.F, Nohaile, M.J, Amy, N.K, Kustu, S, Wemmer, D.E. | | Deposit date: | 1994-09-16 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the N-terminal receiver domain of NTRC.
Biochemistry, 34, 1995
|
|
1XHE
 
 | |
1XHF
 
 | | Crystal structure of the bef3-activated receiver domain of redox response regulator arca | | Descriptor: | Aerobic respiration control protein arcA, BERYLLIUM DIFLUORIDE, BERYLLIUM TETRAFLUORIDE ION, ... | | Authors: | Toro-Roman, A, Mack, T.R, Stock, A.M. | | Deposit date: | 2004-09-18 | | Release date: | 2005-05-17 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structural Analysis and Solution Studies of the Activated Regulatory Domain of the Response Regulator ArcA: A Symmetric Dimer Mediated by the alpha4-beta5-alpha5 Face
J.Mol.Biol., 349, 2005
|
|
1ZGZ
 
 | |
1NXO
 
 | | MicArec pH7.0 | | Descriptor: | DNA-binding response regulator | | Authors: | Bent, C.J, Isaacs, N.W, Mitchell, T.J, Riboldi-Tunnicliffe, A. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states.
J.Bacteriol., 186, 2004
|
|
1NXX
 
 | |
1ZH2
 
 | |
1ZIT
 
 | |
1NXW
 
 | | MicArec pH 5.1 | | Descriptor: | ACETIC ACID, DNA-binding response regulator | | Authors: | Bent, C.J, Isaacs, N.W, Mitchell, T.J, Riboldi-Tunnicliffe, A. | | Deposit date: | 2003-02-11 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of the response regulator 02 receiver domain, the essential YycF two-component system of Streptococcus pneumoniae in both complexed and native states.
J.Bacteriol., 186, 2004
|
|
1ZY2
 
 | | Crystal structure of the phosphorylated receiver domain of the transcription regulator NtrC1 from Aquifex aeolicus | | Descriptor: | MAGNESIUM ION, transcriptional regulator NtrC1 | | Authors: | Doucleff, M, Chen, B, Maris, A.E, Wemmer, D.E, Kondrashkina, E, Nixon, B.T. | | Deposit date: | 2005-06-09 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Negative regulation of AAA + ATPase assembly by two component receiver domains: a transcription activation mechanism that is conserved in mesophilic and extremely hyperthermophilic bacteria
J.Mol.Biol., 353, 2005
|
|
