2BUT
 
 | | Crystal Structure Of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R457S - APO | | Descriptor: | FE (III) ION, HYDROXIDE ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biophysical Analyses of Designed and Selected Mutants of Protocatechuate 3,4-Dioxygenase
Annu.Rev.Microbiol., 58, 2004
|
|
2BUY
 
 | | Crystal Structure of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R133H in Complex with Catechol | | Descriptor: | CATECHOL, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-20 | | Release date: | 2006-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies of Intradiol Dioxygenases.
Phd Thesis, 2001
|
|
1BHH
 
 | | FREE P56LCK SH2 DOMAIN | | Descriptor: | P56 LCK TYROSINE KINASE SH2 DOMAIN, T-LYMPHOCYTE-SPECIFIC PROTEIN TYROSINE KINASE P56LCK | | Authors: | Tong, L, Warren, T.C, Lukas, S, Schembri-King, J, Betageri, R, Proudfoot, J.R, Jakes, S. | | Deposit date: | 1998-06-08 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carboxymethyl-phenylalanine as a replacement for phosphotyrosine in SH2 domain binding.
J.Biol.Chem., 273, 1998
|
|
1BE0
 
 | | HALOALKANE DEHALOGENASE AT PH 5.0 CONTAINING ACETIC ACID | | Descriptor: | ACETATE ION, ACETIC ACID, HALOALKANE DEHALOGENASE | | Authors: | Ridder, I.S, Vos, G.J, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinetic analysis and X-ray structure of haloalkane dehalogenase with a modified halide-binding site.
Biochemistry, 37, 1998
|
|
1BIN
 
 | | LEGHEMOGLOBIN A (ACETOMET) | | Descriptor: | ACETATE ION, LEGHEMOGLOBIN A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brucker, E.A, Hargrove, M.S, Phillips Jr, G.N. | | Deposit date: | 1996-08-23 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants.
J.Mol.Biol., 266, 1997
|
|
2BXM
 
 | | Human serum albumin complexed with myristate and indomethacin | | Descriptor: | INDOMETHACIN, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
1BF4
 
 | | CHROMOSOMAL DNA-BINDING PROTEIN SSO7D/D(GCGAACGC) COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*5IUP*CP*GP*C)-3'), PROTEIN (CHROMOSOMAL PROTEIN SSO7D) | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Padmanabhan, S, Lim, L, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1998-05-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.
Nat.Struct.Biol., 5, 1998
|
|
1BFB
 
 | | BASIC FIBROBLAST GROWTH FACTOR COMPLEXED WITH HEPARIN TETRAMER FRAGMENT | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Faham, S, Rees, D.C. | | Deposit date: | 1995-12-12 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Heparin structure and interactions with basic fibroblast growth factor.
Science, 271, 1996
|
|
1BFO
 
 | |
2BYX
 
 | |
1BGW
 
 | | TOPOISOMERASE RESIDUES 410-1202, | | Descriptor: | TOPOISOMERASE | | Authors: | Berger, J.M, Gamblin, S.J, Harrison, S.C, Wang, J.C. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of DNA topoisomerase II.
Nature, 379, 1996
|
|
2BV0
 
 | | Crystal Structure of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R133H in Complex with Protocatechuate. | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-20 | | Release date: | 2006-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies of Intradiol Dioxygenases.
Phd Thesis, 2001
|
|
1BGT
 
 | |
1BH2
 
 | | A326S MUTANT OF AN INHIBITORY ALPHA SUBUNIT | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GUANINE NUCLEOTIDE-BINDING PROTEIN, MAGNESIUM ION | | Authors: | Mixon, M.B, Posner, B.A, Wall, M.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1998-06-12 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The A326S mutant of Gialpha1 as an approximation of the receptor-bound state.
J.Biol.Chem., 273, 1998
|
|
1BIL
 
 | | CRYSTALLOGRAPHIC STUDIES ON THE BINDING MODES OF P2-P3 BUTANEDIAMIDE RENIN INHIBITORS | | Descriptor: | (2S)-2-[(2-amino-1,3-thiazol-4-yl)methyl]-N~1~-[(1S,2R,3R)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methylhexyl]-N~4~-[2-(d imethylamino)-2-oxoethyl]-N~4~-[(1S)-1-phenylethyl]butanediamide, Renin | | Authors: | Tong, L. | | Deposit date: | 1995-09-27 | | Release date: | 1996-01-29 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies on the binding modes of P2-P3 butanediamide renin inhibitors.
J.Biol.Chem., 270, 1995
|
|
2BUZ
 
 | | Crystal Structure of Protocatechuate 3,4-Dioxygenase from Acinetobacter Sp. ADP1 Mutant R133H in Complex with 4-Nitrocatechol | | Descriptor: | 4-NITROCATECHOL, FE (III) ION, PROTOCATECHUATE 3,4-DIOXYGENASE ALPHA CHAIN, ... | | Authors: | Vetting, M.W, Valley, M.P, D'Argenio, D.A, Ornston, L.N, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2005-06-20 | | Release date: | 2006-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Studies of Intradiol Dioxygenases
Ph D Thesis, 2001
|
|
2C35
 
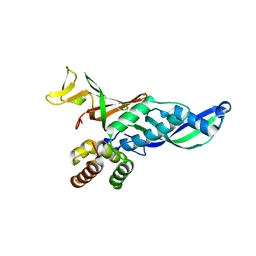 | | Subunits Rpb4 and Rpb7 of human RNA polymerase II | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 16 KDA POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 19 KDA POLYPEPTIDE | | Authors: | Meka, H, Werner, F, Cordell, S.C, Onesti, S, Brick, P. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and RNA Binding of the Rpb4/Rpb7 Subunits of Human RNA Polymerase II.
Nucleic Acids Res., 33, 2005
|
|
2BXQ
 
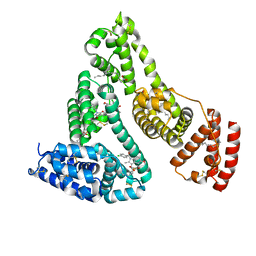 | | Human serum albumin complexed with myristate, phenylbutazone and indomethacin | | Descriptor: | 4-BUTYL-1,2-DIPHENYL-PYRAZOLIDINE-3,5-DIONE, INDOMETHACIN, MYRISTIC ACID, ... | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
2BMK
 
 | |
1BEC
 
 | |
2BQR
 
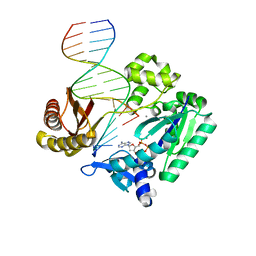 | | DNA Adduct Bypass Polymerization by Sulfolobus solfataricus Dpo4. Analysis and Crystal Structures of Multiple Base-Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*C)-3', 5'-D(*TP*CP*AP*TP*GNEP*GP*AP*AP*TP*CP*CP *TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2005-04-27 | | Release date: | 2005-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | DNA Adduct Bypass Polymerization by Sulfolobus Solfataricus DNA Polymerase Dpo4: Analysis and Crystal Structures of Multiple Base Pair Substitution and Frameshift Products with the Adduct 1,N2-Ethenoguanine.
J.Biol.Chem., 280, 2005
|
|
1BI2
 
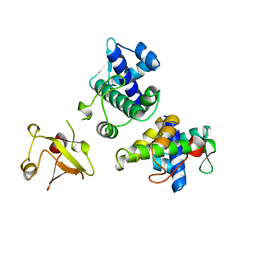 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR | | Authors: | Pohl, E, Hol, W.G.J. | | Deposit date: | 1998-06-21 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
2BSA
 
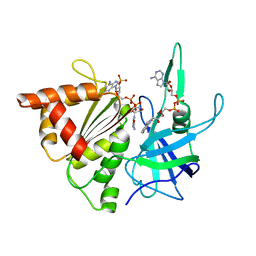 | | Ferredoxin-Nadp Reductase (Mutation: Y 303 S) complexed with NADP | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maya, C.M, Hermoso, J.A, Perez-Dorado, I, Tejero, J, Julvez, M.M, Gomez-Moreno, C, Medina, M. | | Deposit date: | 2005-05-20 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
1BG8
 
 | |
1BGG
 
 | | GLUCOSIDASE A FROM BACILLUS POLYMYXA COMPLEXED WITH GLUCONATE | | Descriptor: | BETA-GLUCOSIDASE A, D-gluconic acid | | Authors: | Sanz-Aparicio, J, Hermoso, J, Martinez-Ripoll, M, Polaina, J. | | Deposit date: | 1997-05-12 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
