7MRI
 
 | | Crystal structure of N63T yeast iso-1-cytochrome c | | Descriptor: | Cytochrome c isoform 1, HEME C | | Authors: | Lei, H, Bowler, B.E, Evenson, G.E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Effect on intrinsic peroxidase activity of substituting coevolved residues from Omega-loop C of human cytochrome c into yeast iso-1-cytochrome c.
J.Inorg.Biochem., 232, 2022
|
|
3AFG
 
 | | Crystal structure of ProN-Tk-SP from Thermococcus kodakaraensis | | Descriptor: | CALCIUM ION, Subtilisin-like serine protease | | Authors: | Foophow, T, Tanaka, S, Angkawidjaja, C, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a subtilisin homologue, Tk-SP, from Thermococcus kodakaraensis: requirement of a C-terminal beta-jelly roll domain for hyperstability.
J.Mol.Biol., 400, 2010
|
|
4AZA
 
 | | Improved eIF4E binding peptides by phage display guided design. | | Descriptor: | EIF4G1_D5S PEPTIDE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Chew, W.Z, Quah, S.T, Verma, C.S, Liu, Y, Lane, D.P, Brown, C.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Improved Eif4E Binding Peptides by Phage Display Guided Design: Plasticity of Interacting Surfaces Yield Collective Effects.
Plos One, 7, 2012
|
|
3IQV
 
 | |
2YK3
 
 | | CRITHIDIA FASCICULATA CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, SULFATE ION | | Authors: | Fulop, V, Sam, K.A, Ferguson, S.J, Ginger, M.L, Allen, J.W.A. | | Deposit date: | 2011-05-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Trypanosomatid Mitochondrial Cytochrome C with Heme Attached Via Only One Thioether Bond and Implications for the Substrate Recognition Requirements of Heme Lyase.
FEBS J., 276, 2009
|
|
3N89
 
 | | KH domains | | Descriptor: | Defective in germ line development protein 3, isoform a | | Authors: | Nakel, K, Hartung, S.A, Bonneau, F, Eckmann, C.R, Conti, E. | | Deposit date: | 2010-05-28 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Four KH domains of the C. elegans Bicaudal-C ortholog GLD-3 form a globular structural platform.
Rna, 16, 2010
|
|
1MG7
 
 | | Crystal Structure of xol-1 | | Descriptor: | early switch protein xol-1 2.2k splice form | | Authors: | Luz, J.G, Hassig, C.A, Godzik, A, Meyer, B.J, Wilson, I.A. | | Deposit date: | 2002-08-14 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators
Genes Dev., 17, 2003
|
|
1YDK
 
 | | Crystal structure of the I219A mutant of human glutathione transferase A1-1 with S-hexylglutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Kuhnert, D.C, Sayed, Y, Mosebi, S, Sayed, M, Sewell, T, Dirr, H.W. | | Deposit date: | 2004-12-24 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Interactions Stabilise the C-terminal Region of Human Glutathione Transferase A1-1: a Crystallographic and Calorimetric Study.
J.Mol.Biol., 349, 2005
|
|
3N97
 
 | | RNA polymerase alpha C-terminal domain (E. coli) and sigma region 4 (T. aq. mutant) bound to (UP,-35 element) DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*CP*CP*AP*TP*GP*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*TP*TP*TP*TP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*AP*AP*AP*AP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Lara-Gonzalez, S, Birktoft, J.J, Lawson, C.L. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.252 Å) | | Cite: | The RNA Polymerase alpha Subunit Recognizes the DNA Shape of the Upstream Promoter Element.
Biochemistry, 59, 2020
|
|
4JHS
 
 | | Crystal structure of a C-terminal two domain fragment of human beta-2-glycoprotein 1 | | Descriptor: | Beta-2-glycoprotein 1, SULFATE ION | | Authors: | Bonanno, J.B, Toro, R, Gizzi, A, Chan, M.K, Garrett-Thomson, S.C, Patel, H, Lim, S, Matikainen, B, Celikgil, A, Garforth, S, Hillerich, B, Seidel, R, Rand, J.H, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a C-terminal two domain fragment of human beta-2-glycoprotein 1
To be Published
|
|
1OS6
 
 | | Cytochrome c7 (PpcA) from Geobacter sulfurreducens | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, HEME C, PpcA, ... | | Authors: | Pokkuluri, P.R, Londer, Y.Y, Duke, N.E.C, Long, W.C, Schiffer, M. | | Deposit date: | 2003-03-18 | | Release date: | 2004-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Family of cytochrome c7-type proteins from Geobacter sulfurreducens: structure of one cytochrome c7 at 1.45 A resolution.
Biochemistry, 43, 2004
|
|
3QIU
 
 | | Crystal structure of the 226 TCR in complex with MCC/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, E-K alpha chain, ... | | Authors: | Kruse, A.C, Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3D5M
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-({3-[(5S)-5-tert-butyl-1-(3-chloro-4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzis othiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-05-16 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of 1,1-dioxoisothiazole and benzo[b]thiophene-1,1-dioxide derivatives as novel inhibitors of hepatitis C virus NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1KX2
 
 | | Minimized average structure of a mono-heme ferrocytochrome c from Shewanella putrefaciens | | Descriptor: | HEME C, mono-heme c-type cytochrome ScyA | | Authors: | Bartalesi, I, Bertini, I, Hajieva, P, Rosato, A, Vasos, P.R. | | Deposit date: | 2002-01-30 | | Release date: | 2002-02-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a monoheme ferrocytochrome c from Shewanella putrefaciens and structural analysis of sequence-similar proteins: functional implications.
Biochemistry, 41, 2002
|
|
1KX7
 
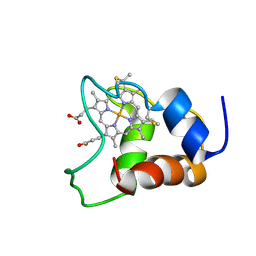 | | Family of 30 conformers of a mono-heme ferrocytochrome c from Shewanella putrefaciens solved by NMR | | Descriptor: | HEME C, mono-heme c-type cytochrome ScyA | | Authors: | Bartalesi, I, Bertini, I, Hajieva, P, Rosato, A, Vasos, P.R. | | Deposit date: | 2002-01-31 | | Release date: | 2002-02-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a monoheme ferrocytochrome c from Shewanella putrefaciens and structural analysis of sequence-similar proteins: functional implications.
Biochemistry, 41, 2002
|
|
3D28
 
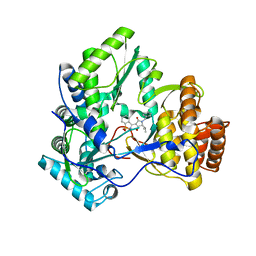 | | Crystal structure of hcv ns5b polymerase with a novel benzisothiazole inhibitor | | Descriptor: | (5S)-1-benzyl-3-(1,1-dioxido-1,2-benzisothiazol-3-yl)-4-hydroxy-5-(1-methylethyl)-1,5-dihydro-2H-pyrrol-2-one, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2008-05-07 | | Release date: | 2009-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of 1,1-dioxoisothiazole and benzo[b]thiophene-1,1-dioxide derivatives as novel inhibitors of hepatitis C virus NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1U5M
 
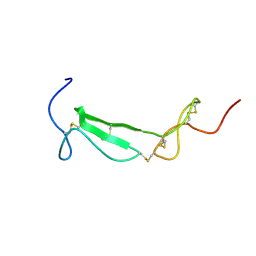 | | Structure of a Chordin-like Cysteine-rich Repeat (VWC module) from Collagen IIA | | Descriptor: | alpha 1 type II collagen isoform 1 | | Authors: | O'Leary, J.M, Hamilton, J.m, Deane, C.M, Valeyev, N.V, Sandell, L.J, Downing, A.K. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-05 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA
J.Biol.Chem., 279, 2004
|
|
1D4D
 
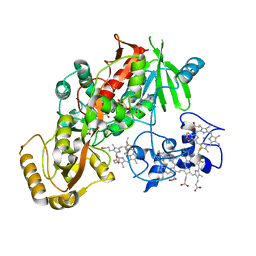 | | CRYSTAL STRUCTURE OF THE SUCCINATE COMPLEXED FORM OF THE FLAVOCYTOCHROME C FUMARATE REDUCTASE OF SHEWANELLA PUTREFACIENS STRAIN MR-1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C FUMARATE REDUCTASE, HEME C, ... | | Authors: | Leys, D, Tsapin, A.S, Meyer, T.E, Cusanovich, M.A, Van Beeumen, J.J. | | Deposit date: | 1999-10-03 | | Release date: | 1999-12-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the flavocytochrome c fumarate reductase of Shewanella putrefaciens MR-1.
Nat.Struct.Biol., 6, 1999
|
|
5MVO
 
 | | FoxE P43212 crystal structure of Rhodopseudomonas ferrooxidans SW2 putative iron oxidase | | Descriptor: | COPPER (II) ION, FoxE, HEME C, ... | | Authors: | Pereira, L, Saraiva, I.H, Oliveira, A.S, Soares, C, Gomes, R.O, Frazao, C. | | Deposit date: | 2017-01-17 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.668 Å) | | Cite: | Crystallization and preliminary crystallographic studies of FoxE from Rhodobacter ferrooxidans SW2, an Fe(II) oxidoreductase involved in photoferrotrophy.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 68, 2012
|
|
1D0H
 
 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH N-ACETYL-GALACTOSAMINE | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, PROTEIN (TETANUS TOXIN HC), SULFATE ION | | Authors: | Emsley, P, Fotinou, C, Black, I, Fairweather, N.F, Charles, I.G, Watts, C, Hewitt, E, Isaacs, N.W. | | Deposit date: | 1999-09-10 | | Release date: | 2000-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding.
J.Biol.Chem., 275, 2000
|
|
1DFQ
 
 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH SIALIC ACID | | Descriptor: | N-acetyl-beta-neuraminic acid, TETANUS TOXIN HC | | Authors: | Emsley, P, Fotinou, C, Black, I, Fairweather, N.F, Charles, I.G, Watts, C, Hewitt, E, Isaacs, N.W. | | Deposit date: | 1999-11-20 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding.
J.Biol.Chem., 275, 2000
|
|
4Q96
 
 | | CID of human RPRD1B in complex with an unmodified CTD peptide | | Descriptor: | RPB1-CTD, Regulation of nuclear pre-mRNA domain-containing protein 1B, SULFATE ION, ... | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-29 | | Release date: | 2014-06-04 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3ZYU
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with I-BET151(GSK1210151A) | | Descriptor: | 1,2-ETHANEDIOL, 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C.W, Mirguet, O. | | Deposit date: | 2011-08-25 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibition of Bet Recruitment to Chromatin as an Effective Treatment for Mll-Fusion Leukaemia.
Nature, 478, 2011
|
|
4E93
 
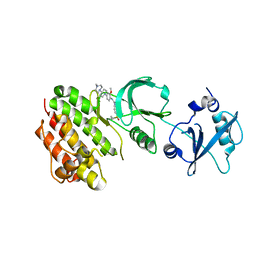 | | Crystal structure of human Feline Sarcoma Viral Oncogene Homologue (v-FES)in complex with TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, Tyrosine-protein kinase Fes/Fps | | Authors: | Filippakopoulos, P, Salah, E, Miduturu, C.V, Fedorov, O, Cooper, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Small-Molecule Inhibitors of the c-Fes Protein-Tyrosine Kinase.
Chem.Biol., 19, 2012
|
|
4AEJ
 
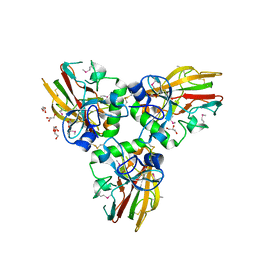 | | Crystal structure of Human fibrillar procollagen type III C- propeptide trimer | | Descriptor: | CALCIUM ION, COLLAGEN ALPHA-1(III) CHAIN, GLYCEROL | | Authors: | Bourhis, J.M, Mariano, N, Zhao, Y, Harlos, K, Jones, E.Y, Moali, C, Aghajari, N, Hulmes, D.J.S. | | Deposit date: | 2012-01-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis of Fibrillar Collagen Trimerization and Related Genetic Disorders.
Nat.Struct.Mol.Biol., 19, 2012
|
|
