3OY9
 
 | |
1AJ1
 
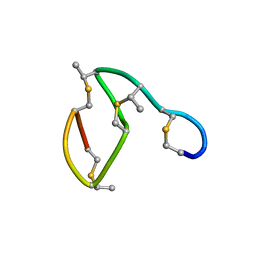 | | NMR STRUCTURE OF THE LANTIBIOTIC ACTAGARDINE | | Descriptor: | LANTIBIOTIC ACTAGARDINE | | Authors: | Zimmermann, N, Jung, G. | | Deposit date: | 1997-05-14 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the lantibiotic murein-biosynthesis-inhibitor actagardine determined by NMR.
Eur.J.Biochem., 246, 1997
|
|
4BT8
 
 | |
198D
 
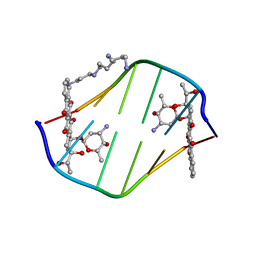 | | A TRIGONAL FORM OF THE IDARUBICIN-D(CGATCG) COMPLEX: CRYSTAL AND MOLECULAR STRUCTURE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), IDARUBICIN, SPERMINE | | Authors: | Dautant, A, Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | Deposit date: | 1994-11-28 | | Release date: | 1995-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A trigonal form of the idarubicin:d(CGATCG) complex; crystal and molecular structure at 2.0 A resolution.
Nucleic Acids Res., 23, 1995
|
|
4BTA
 
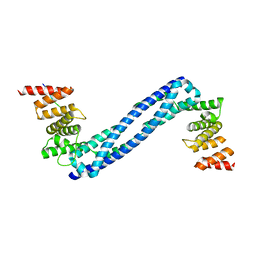 | | CRYSTAL STRUCTURE OF THE PEPTIDE(PRO-PRO-GLY)3 BOUND COMPLEX OF N- TERMINAL DOMAIN AND PEPTIDE SUBSTRATE BINDING DOMAIN OF PROLYL-4 HYDROXYLASE (RESIDUES 1-244) TYPE I FROM HUMAN | | Descriptor: | PROLINE RICH PEPTIDE, PROLYL 4-HYDROXYLASE SUBUNIT ALPHA-1 | | Authors: | Anantharajan, J, Koski, M.K, Pekkala, M, Wierenga, R.K. | | Deposit date: | 2013-06-14 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The Structural Motifs for Substrate Binding and Dimerization of the Alpha Subunit of Collagen Prolyl 4-Hydroxylase
Structure, 21, 2013
|
|
1ALN
 
 | |
1A8I
 
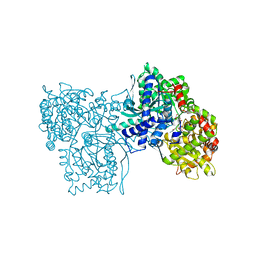 | | SPIROHYDANTOIN INHIBITOR OF GLYCOGEN PHOSPHORYLASE | | Descriptor: | BETA-D-GLUCOPYRANOSE SPIROHYDANTOIN, GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-25 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
4JJ7
 
 | |
1ADZ
 
 | |
1B58
 
 | |
4O5J
 
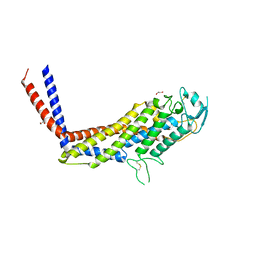 | | Crystal structure of SabA from Helicobacter pylori | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Uncharacterized protein | | Authors: | Pang, S.S, Nguyen, S.T.S, Whisstock, J.C. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-01 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure of the extracellular adhesion domain of the sialic acid-binding adhesin SabA from Helicobacter pylori
J.Biol.Chem., 289, 2013
|
|
3E76
 
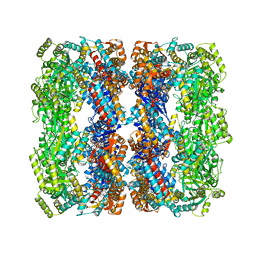 | | Crystal structure of Wild-type GroEL with bound Thallium ions | | Descriptor: | 60 kDa chaperonin, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kiser, P.D, Lorimer, G.H, Palczewski, K. | | Deposit date: | 2008-08-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Use of thallium to identify monovalent cation binding sites in GroEL.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
4J8F
 
 | |
1AQ7
 
 | |
4AJB
 
 | | 3D structure of E. coli Isocitrate Dehydrogenase K100M mutant in complex with Isocitrate, magnesium(II) and thioNADP | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ISOCITRIC ACID, MAGNESIUM ION, ... | | Authors: | Goncalves, S, Miller, S.P, Carrondo, M.A, Dean, A.M, Matias, P.M. | | Deposit date: | 2012-02-16 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Induced Fit and the Catalytic Mechanism of Isocitrate Dehydrogenase.
Biochemistry, 51, 2012
|
|
1DXQ
 
 | | CRYSTAL STRUCTURE OF MOUSE NAD[P]H-QUINONE OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, QUINONE REDUCTASE | | Authors: | Faig, M, Bianchet, M.A, Chen, S, Winski, S, Ross, D, Amzel, L.M. | | Deposit date: | 2000-01-14 | | Release date: | 2000-04-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Recombinant Mouse and Human Nad(P)H:Quinone Oxidoreductases:Species Comparison and Structural Changes with Substrate Binding and Release
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4AEY
 
 | |
3CHF
 
 | |
2XQ4
 
 | | Pentameric ligand gated ion channel GLIC in complex with tetramethylarsonium (TMAs) | | Descriptor: | ARSENIC, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
1B5J
 
 | |
1B6H
 
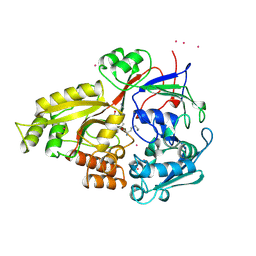 | |
3D1D
 
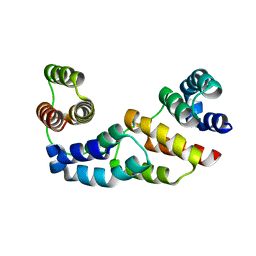 | | Hexagonal crystal structure of Tas3 C-terminal alpha motif | | Descriptor: | RNA-induced transcriptional silencing complex protein tas3 | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing.
Mol.Cell, 34, 2009
|
|
1B7H
 
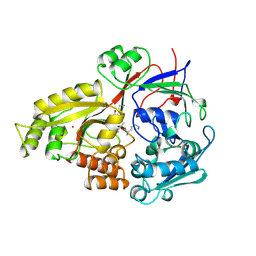 | |
4KNE
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
3PRK
 
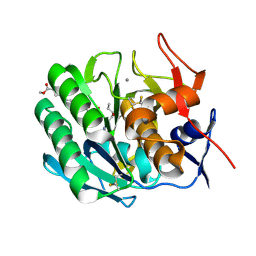 | | INHIBITION OF PROTEINASE K BY METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE. AN X-RAY STUDY AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE, PROTEINASE K | | Authors: | Wolf, W.M, Bajorath, J, Mueller, A, Raghunathan, S, Singh, T.P, Hinrichs, W, Saenger, W. | | Deposit date: | 1991-08-07 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of proteinase K by methoxysuccinyl-Ala-Ala-Pro-Ala-chloromethyl ketone. An x-ray study at 2.2-A resolution.
J.Biol.Chem., 266, 1991
|
|
