9FGQ
 
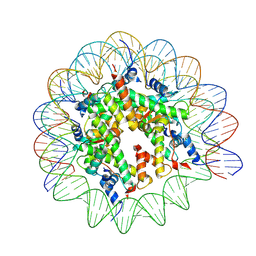 | | Structure of human APC3loop 375-381 bound to the NCP | | Descriptor: | Cell division cycle protein 27 homolog, DNA (131-MER), DNA (132-MER), ... | | Authors: | Young, R.V.C, Muhammad, R, Alfieri, C. | | Deposit date: | 2024-05-24 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Spatial control of the APC/C ensures the rapid degradation of cyclin B1.
Embo J., 43, 2024
|
|
9FEH
 
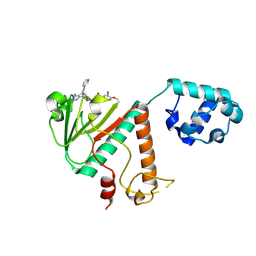 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the STM957 inhibitor | | Descriptor: | Transcription factor ETV6,Guanine-N7 methyltransferase nsp14, ZINC ION, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-pyridin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-cyano-~{N}-ethyl-4-methoxy-benzenesulfonamide | | Authors: | Zilecka, E, Klima, M, Boura, E. | | Deposit date: | 2024-05-20 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of SARS-CoV-2 MTase nsp14 with the inhibitor STM957 reveals inhibition mechanism that is shared with a poxviral MTase VP39.
J Struct Biol X, 10, 2024
|
|
9FEF
 
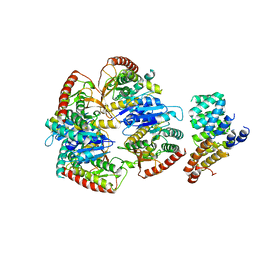 | | Cryo-EM structure of Trypanosoma cruzi (MDH)4-PEX5 complex | | Descriptor: | Peroxisome targeting signal 1 receptor, malate dehydrogenase | | Authors: | Lipinski, O, Sonani, R.R, Blat, A, Jemiola-Rzeminska, M, Patel, S.N, Sood, T, Dubin, G. | | Deposit date: | 2024-05-19 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Cryo-EM structure of Trypanosoma cruzi (MDH)4-PEX5 complex
To Be Published
|
|
9FEA
 
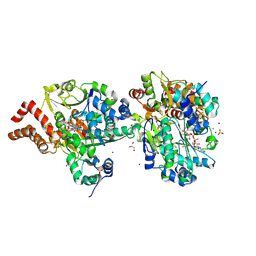 | | Crystal Structure of reduced NuoEF variant P228R(NuoF) from Aquifex aeolicus bound to NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FE8
 
 | | Crystal Structure of reduced NuoEF variant P228R(NuoF) from Aquifex aeolicus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FE7
 
 | | Crystal Structure of oxidized NuoEF variant P228R(NuoF) from Aquifex aeolicus | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FE5
 
 | | Crystal Structure of NuoEF variant R66G(NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions after 10 min soaking | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FE0
 
 | | Crystal Structure of reduced NuoEF variant R66G(NuoF) from Aquifex aeolicus bound to NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FDV
 
 | | Crystal Structure of reduced NuoEF variant R66G(NuoF) from Aquifex aeolicus | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FDK
 
 | | Crystal Structure of oxidized NuoEF variant R66G(NuoF) from Aquifex aeolicus | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FDJ
 
 | | Crystal structure of the NuoEF variant R66G (NuoF) from Aquifex aeolicus bound to NADH under anoxic conditions (short soak) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Wohlwend, D, Friedrich, T, Goeppert-Asadollahpour, S. | | Deposit date: | 2024-05-17 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural robustness of the NADH binding site in NADH:ubiquinone oxidoreductase (complex I).
Biochim Biophys Acta Bioenerg, 1865, 2024
|
|
9FCR
 
 | |
9FCG
 
 | | Medicago truncatula 5'-ProFAR isomerase (HISN3) D57N mutant in complex with PrFAR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic, ... | | Authors: | Witek, W, Imiolczyk, B, Ruszkowski, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural, kinetic, and evolutionary peculiarities of HISN3, a plant 5'-ProFAR isomerase.
Plant Physiol Biochem., 215, 2024
|
|
9FCF
 
 | | Medicago truncatula 5'-ProFAR isomerase (HISN3) D57N mutant in complex with ProFAR | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, chloroplastic, CHLORIDE ION, ... | | Authors: | Witek, W, Imiolczyk, B, Ruszkowski, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural, kinetic, and evolutionary peculiarities of HISN3, a plant 5'-ProFAR isomerase.
Plant Physiol Biochem., 215, 2024
|
|
9FBF
 
 | | VDR complex with UG-481 | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(1~{S})-5-methyl-1-[(1~{R},2~{R})-2-(3-methyl-3-oxidanyl-butyl)cyclopropyl]-5-oxidanyl-hexyl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2024-05-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of New Type of Gemini Analogues with a Cyclopropane Moiety in Their Side Chain.
J.Med.Chem., 67, 2024
|
|
9F9U
 
 | |
9F9Q
 
 | |
9F7X
 
 | |
9F7W
 
 | |
9F7H
 
 | | UP1 in complex with Z45617795 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, N-(2-phenylethyl)methanesulfonamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F7F
 
 | | UP1 in complex with Z1491353358 | | Descriptor: | (3S)-1-(phenylsulfonyl)pyrrolidin-3-amine, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-05-03 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
9F6L
 
 | |
9F6K
 
 | |
9F6J
 
 | |
9F6I
 
 | |
