1WLH
 
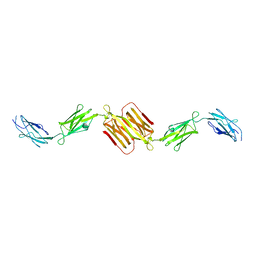 | | Molecular structure of the rod domain of Dictyostelium filamin | | Descriptor: | Gelation factor | | Authors: | Popowicz, G.M, Mueller, R, Noegel, A.A, Schleicher, M, Huber, R, Holak, T.A. | | Deposit date: | 2004-06-27 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of the rod domain of dictyostelium filamin
J.Mol.Biol., 342, 2004
|
|
1WVZ
 
 | |
1WR6
 
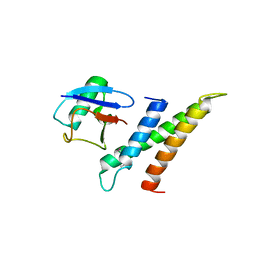 | | Crystal structure of GGA3 GAT domain in complex with ubiquitin | | Descriptor: | ADP-ribosylation factor binding protein GGA3, ubiquitin | | Authors: | Kawasaki, M, Shiba, T, Shiba, Y, Yamaguchi, Y, Matsugaki, N, Igarashi, N, Suzuki, M, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-12 | | Release date: | 2005-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of ubiquitin recognition by GGA3 GAT domain.
Genes Cells, 10, 2005
|
|
1WRD
 
 | | Crystal structure of Tom1 GAT domain in complex with ubiquitin | | Descriptor: | Target of Myb protein 1, Ubiquitin | | Authors: | Akutsu, M, Kawasaki, M, Katoh, Y, Shiba, T, Yamaguchi, Y, Kato, R, Kato, K, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2004-10-14 | | Release date: | 2005-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for recognition of ubiquitinated cargo by Tom1-GAT domain.
Febs Lett., 579, 2005
|
|
1WUW
 
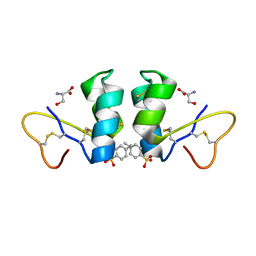 | | Crystal Structure of beta hordothionin | | Descriptor: | Beta-hordothionin, PARA-TOLUENE SULFONATE, SERINE | | Authors: | Johnson, K.A, Kim, E, Teeter, M.M, Suh, S.W, Stec, B. | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alpha-hordothionin at 1.9 Angstrom resolution.
Febs Lett., 579, 2005
|
|
1XEE
 
 | | Solution structure of the Chemotaxis Inhibitory Protein of Staphylococcus aureus | | Descriptor: | chemotaxis-inhibiting protein CHIPS | | Authors: | Haas, P.J, de Haas, C.J, Poppelier, M.J, van Kessel, K.P, van Strijp, J.A, Dijkstra, K, Scheek, R.M, Fan, H, Kruijtzer, J.A, Liskamp, R.M, Kemmink, J. | | Deposit date: | 2004-09-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the C5a receptor-blocking domain of chemotaxis inhibitory protein of Staphylococcus aureus is related to a group of immune evasive molecules
J.Mol.Biol., 353, 2005
|
|
1X6W
 
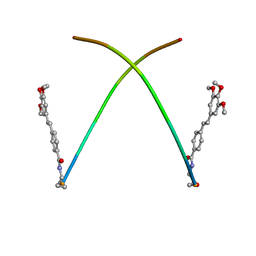 | |
1XUB
 
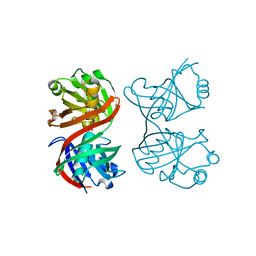 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
1XU6
 
 | | Structure of the C-terminal domain from Trypanosoma brucei Variant Surface Glycoprotein MITat1.2 | | Descriptor: | Variant surface glycoprotein MITAT 1.2 | | Authors: | Chattopadhyay, A, Jones, N.G, Nietlispach, D, Nielsen, P.R, Voorheis, H.P, Mott, H.R, Carrington, M. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain from Trypanosoma brucei variant surface glycoprotein MITat1.2
J.Biol.Chem., 280, 2004
|
|
1YJB
 
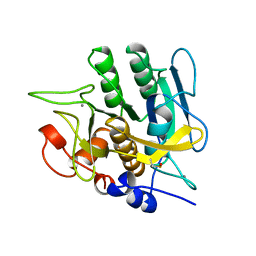 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 35% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YUR
 
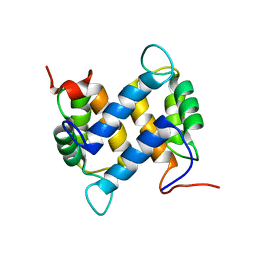 | | Solution structure of apo-S100A13 (minimized mean structure) | | Descriptor: | S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YP8
 
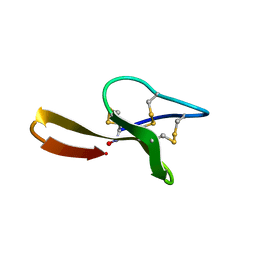 | |
1YP6
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1YJA
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 20% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YJC
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 50% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YUW
 
 | |
3MU6
 
 | | Inhibiting the Binding of Class IIa Histone Deacetylases to Myocyte Enhancer Factor-2 by Small Molecules | | Descriptor: | (3E)-N~8~-(2-aminophenyl)-N~1~-phenyloct-3-enediamide, DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), ... | | Authors: | Jayathilaka, N, Han, A, Gaffney, K, Dey, R, He, J, Ye, J, Gao, T, Petasis, N.A, Chen, L. | | Deposit date: | 2010-05-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.434 Å) | | Cite: | Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2.
Nucleic Acids Res., 40, 2012
|
|
1YIK
 
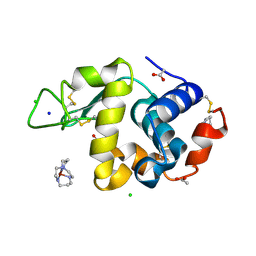 | | Structure of Hen egg white lysozyme soaked with Cu-cyclam | | Descriptor: | 1,4,8,11-TETRAAZA-CYCLOTETRADECANE CU(II), ACETATE ION, CHLORIDE ION, ... | | Authors: | Hunter, T.M, McNae, I.W, Liang, X, Bella, J, Parsons, S, Walkinshaw, M.D, Sadler, P.J. | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein recognition of macrocycles: binding of anti-HIV metallocyclams to lysozyme
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y62
 
 | | A 2.4 crystal structure of conkunitzin-S1, a novel Kunitz-fold cone snail neurotoxin. | | Descriptor: | Conkunitzin-S1, SULFATE ION | | Authors: | Dy, C.Y, Buczek, P, Horvath, M.P. | | Deposit date: | 2004-12-03 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of conkunitzin-S1, a neurotoxin and Kunitz-fold disulfide variant from cone snail.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1XUA
 
 | | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-10-26 | | Release date: | 2004-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XQ8
 
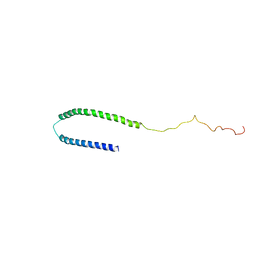 | |
1YUU
 
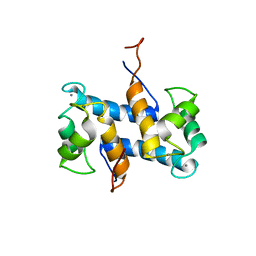 | | Solution structure of Calcium-S100A13 | | Descriptor: | CALCIUM ION, S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YY6
 
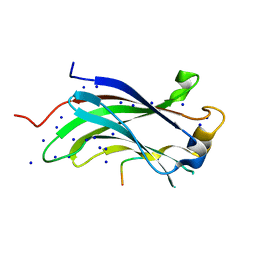 | | The Crystal Structure of the N-terminal domain of HAUSP/USP7 complexed with an EBNA1 peptide | | Descriptor: | Epstein-Barr nuclear antigen-1, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M, Shire, K, Nguyen, T, Zhang, R, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | Deposit date: | 2005-02-23 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
1ZJQ
 
 | |
1Z96
 
 | | Crystal structure of the Mud1 UBA domain | | Descriptor: | UBA-domain protein mud1 | | Authors: | Trempe, J.-F, Brown, N.R, Lowe, E.D, Noble, M.E.M, Gordon, C, Campbell, I.D, Johnson, L.N, Endicott, J.A. | | Deposit date: | 2005-03-31 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain
Embo J., 24, 2005
|
|
