1HUH
 
 | |
1LIE
 
 | |
1HOC
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF H-2DB AT 2.4 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ANTIGEN-DETERMINANT SELECTION | | Descriptor: | 9-RESIDUE PEPTIDE, BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (H2-DB) (ALPHA CHAIN) | | Authors: | Young, A.C.M, Zhang, W, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1994-04-30 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structure of H-2Db at 2.4 A resolution: implications for antigen-determinant selection
Cell(Cambridge,Mass.), 76, 1994
|
|
1NNA
 
 | | THREE-DIMENSIONAL STRUCTURE OF INFLUENZA A N9 NEURAMINIDASE AND ITS COMPLEX WITH THE INHIBITOR 2-DEOXY 2,3-DEHYDRO-N-ACETYL NEURAMINIC ACID | | Descriptor: | CALCIUM ION, NEURAMINIDASE | | Authors: | Bossart-Whitaker, P, Carson, M, Babu, Y.S, Smith, C.D, Laver, W.G, Air, G.M. | | Deposit date: | 1993-03-08 | | Release date: | 1994-04-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of influenza A N9 neuraminidase and its complex with the inhibitor 2-deoxy 2,3-dehydro-N-acetyl neuraminic acid.
J.Mol.Biol., 232, 1993
|
|
1HUG
 
 | |
1HUN
 
 | |
1PYD
 
 | |
1PST
 
 | | CRYSTALLOGRAPHIC ANALYSES OF SITE-DIRECTED MUTANTS OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Chirino, A.J, Feher, G, Rees, D.C. | | Deposit date: | 1993-12-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analyses of site-directed mutants of the photosynthetic reaction center from Rhodobacter sphaeroides.
Biochemistry, 33, 1994
|
|
1PSP
 
 | | PANCREATIC SPASMOLYTIC POLYPEPTIDE: FIRST THREE-DIMENSIONAL STRUCTURE OF A MEMBER OF THE MAMMALIAN TREFOIL FAMILY OF PEPTIDES | | Descriptor: | PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | Gajhede, M, Petersen, T.N, Henriksen, A, Petersen, J.F.W, Dauter, Z, Wilson, K.S, Thim, L. | | Deposit date: | 1994-01-05 | | Release date: | 1994-04-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pancreatic spasmolytic polypeptide: first three-dimensional structure of a member of the mammalian trefoil family of peptides.
Structure, 1, 1993
|
|
1CSH
 
 | |
1CHM
 
 | | ENZYMATIC MECHANISM OF CREATINE AMIDINOHYDROLASE AS DEDUCED FROM CRYSTAL STRUCTURES | | Descriptor: | CARBAMOYL SARCOSINE, CREATINE AMIDINOHYDROLASE | | Authors: | Hoeffken, H.W, Knof, S.H, Bartlett, P.A, Huber, R, Moellering, H, Schumacher, G. | | Deposit date: | 1993-07-19 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzymatic mechanism of creatine amidinohydrolase as deduced from crystal structures.
J.Mol.Biol., 214, 1990
|
|
140D
 
 | | SOLUTION STRUCTURE OF A CONSERVED DNA SEQUENCE FROM THE HIV-1 GENOME: RESTRAINED MOLECULAR DYNAMICS SIMULATION WITH DISTANCE AND TORSION ANGLE RESTRAINTS DERIVED FROM TWO-DIMENSIONAL NMR SPECTRA | | Descriptor: | DNA (5'-D(*AP*GP*CP*TP*TP*GP*CP*CP*TP*TP*GP*AP*G)-3'), DNA (5'-D(*CP*TP*CP*AP*AP*GP*GP*CP*AP*AP*GP*CP*T)-3') | | Authors: | Mujeeb, A, Kerwin, S.M, Kenyon, G.L, James, T.L. | | Deposit date: | 1993-09-24 | | Release date: | 1994-04-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conserved DNA sequence from the HIV-1 genome: restrained molecular dynamics simulation with distance and torsion angle restraints derived from two-dimensional NMR spectra.
Biochemistry, 32, 1993
|
|
142D
 
 | |
141D
 
 | |
143D
 
 | |
136D
 
 | |
1HLC
 
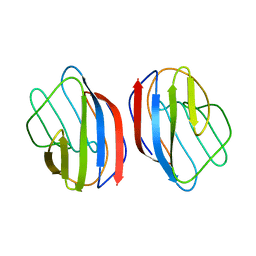 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN DIMERIC S-LAC LECTIN, L-14-II, IN COMPLEX WITH LACTOSE AT 2.9 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN LECTIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Lobsanov, Y.D, Gitt, M.A, Leffler, H, Barondes, S, Rini, J.M. | | Deposit date: | 1993-10-13 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the human dimeric S-Lac lectin, L-14-II, in complex with lactose at 2.9-A resolution.
J.Biol.Chem., 268, 1993
|
|
1ELA
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-prolinamide, ACETIC ACID, CALCIUM ION, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
135D
 
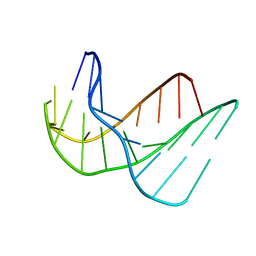 | |
149L
 
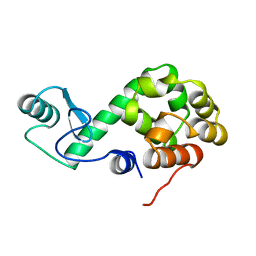 | |
2FGW
 
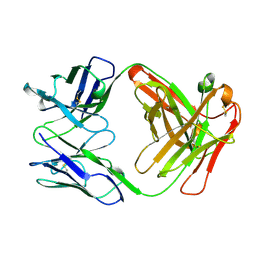 | |
156D
 
 | |
1LIF
 
 | |
1LOG
 
 | |
2TBS
 
 | |
