1LHF
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BORO-HOMOLYS-OH | | Descriptor: | AC-(D)PHE-PRO-BOROHOMOLYS-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
5O52
 
 | | Glycogen Phosphorylase b in complex with 33b | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-5-azanyl-2-(hydroxymethyl)-6-(4-naphthalen-2-yl-1~{H}-imidazol-2-yl)oxane-3,4-diol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, A.L, Kyriakis, E, Stravodimos, G.A, Solovou, T.G.A, Chatzileontiadou, D.S, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanomolar Inhibitors of Glycogen Phosphorylase Based on beta-d-Glucosaminyl Heterocycles: A Combined Synthetic, Enzyme Kinetic, and Protein Crystallography Study.
J. Med. Chem., 60, 2017
|
|
3KGF
 
 | | The structure of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Mycobacterium tuberculosis complexed with phenylalanine and tryptophan | | Descriptor: | CHLORIDE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Webby, C.J, Baker, E.N, Baker, H.M, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2009-10-29 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
5O50
 
 | | Glycogen Phosphorylase b in complex with 33a | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-5-azanyl-2-(hydroxymethyl)-6-(4-phenyl-1~{H}-imidazol-2-yl)oxane-3,4-diol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, A.L, Kyriakis, E, Stravodimos, G.A, Solovou, T.G.A, Chatzileontiadou, D.S, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanomolar Inhibitors of Glycogen Phosphorylase Based on beta-d-Glucosaminyl Heterocycles: A Combined Synthetic, Enzyme Kinetic, and Protein Crystallography Study.
J. Med. Chem., 60, 2017
|
|
5YIF
 
 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, E163A mutant pyruvylated beta-D-galactose complex | | Descriptor: | (2R,4aR,6R,7R,8R,8aR)-2-methyl-6,7,8-tris(oxidanyl)-4,4a,6,7,8,8a-hexahydropyrano[3,2-d][1,3]dioxine-2-carboxylic acid, Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-10-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
2X42
 
 | |
4M7Z
 
 | | Unliganded 1 crystal structure of S25-26 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Haji-Ghassemi, O, Evans, S.V, Muller-Loennies, S, Saldova, R, Muniyappa, M, Brade, L, Rudd, P.M, Harvey, D.J, Kosma, P, Brade, H. | | Deposit date: | 2013-08-12 | | Release date: | 2014-04-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Groove-type Recognition of Chlamydiaceae-specific Lipopolysaccharide Antigen by a Family of Antibodies Possessing an Unusual Variable Heavy Chain N-Linked Glycan.
J.Biol.Chem., 289, 2014
|
|
5O54
 
 | | Glycogen Phosphorylase b in complex with 29a | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-5-azanyl-2-(hydroxymethyl)-6-(5-phenyl-4~{H}-1,2,4-triazol-3-yl)oxane-3,4-diol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Stravodimos, G.A, Kantsadi, A.L, Chatzileontiadou, D.S, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Nanomolar Inhibitors of Glycogen Phosphorylase Based on beta-d-Glucosaminyl Heterocycles: A Combined Synthetic, Enzyme Kinetic, and Protein Crystallography Study.
J. Med. Chem., 60, 2017
|
|
4MOH
 
 | | Pyranose 2-oxidase V546C mutant with 2-fluorinated glucose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-beta-D-glucopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4MOO
 
 | | Pyranose 2-oxidase H450G mutant with 2-fluorinated galactose | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-galactopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
3OUT
 
 | | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate. | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Kudriska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-15 | | Release date: | 2010-09-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of glutamate racemase from Francisella tularensis subsp. tularensis SCHU S4 in complex with D-glutamate.
To be Published
|
|
3TAO
 
 | | Structure of Mycobacterium tuberculosis triosephosphate isomerase bound to phosphoglycolohydroxamate | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, Triosephosphate isomerase | | Authors: | Connor, S.E, Capodagli, G.C, Deaton, M.K, Pegan, S.D. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of Mycobacterium tuberculosis triosephosphate isomerase.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3SS2
 
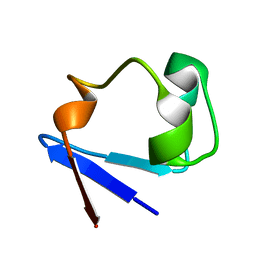 | | Neutron structure of perdeuterated rubredoxin using 48 hours 3rd pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Blakeley, M.P, Weiss, K.L, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4MOG
 
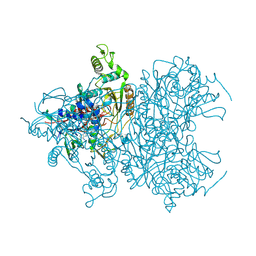 | | Pyranose 2-oxidase V546C mutant with 3-fluorinated glucose | | Descriptor: | 3-deoxy-3-fluoro-beta-D-glucopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4QHV
 
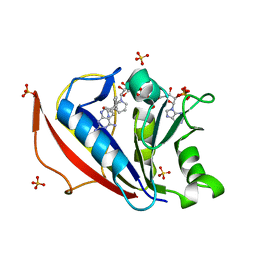 | | Crystal structure of human dihydrofolate reductase as complex with pyridopyrimidine 22 (N~6~-METHYL-N~6~-[4-(PROPAN-2-YL)PHENYL]PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE) | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-[4-(propan-2-yl)phenyl]pyrido[2,3-d]pyrimidine-2,4,6-triamine, ... | | Authors: | Cody, V. | | Deposit date: | 2014-05-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4QJC
 
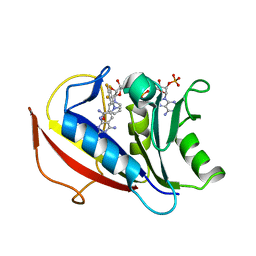 | | Human dihydrofolate reductase ternary complex with NADPH and inhibitor 26 (N~6~-METHYL-N~6~-(3,4,5-TRIFLUOROPHENYL)PYRIDO[2,3-D]PYRIMIDINE-2,4,6-TRIAMINE) | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, N~6~-methyl-N~6~-(3,4,5-trifluorophenyl)pyrido[2,3-d]pyrimidine-2,4,6-triamine | | Authors: | Cody, V. | | Deposit date: | 2014-06-03 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-activity correlations for three pyrido[2,3-d]pyrimidine antifolates binding to human and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
9GCA
 
 | |
4QN9
 
 | | Structure of human NAPE-PLD | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 1,2-Distearoyl-sn-glycerophosphoethanolamine, N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D, ... | | Authors: | Garau, G. | | Deposit date: | 2014-06-17 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Structure of human N-acylphosphatidylethanolamine-hydrolyzing phospholipase D: regulation of fatty acid ethanolamide biosynthesis by bile acids.
Structure, 23, 2015
|
|
9GC8
 
 | | Crystal structure of Pseudomonas aeruginosa IspD in complex with C11H12N2O3 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, methyl 2-[(5~{R})-6-methyl-7-oxidanylidene-5~{H}-pyrrolo[3,4-b]pyridin-5-yl]ethanoate | | Authors: | Borel, F, D'Auria, L. | | Deposit date: | 2024-08-01 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment Discovery by X-Ray Crystallographic Screening Targeting the CTP Binding Site of Pseudomonas Aeruginosa IspD.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
5O56
 
 | | Glycogen Phosphorylase b in complex with 29b | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-5-azanyl-2-(hydroxymethyl)-6-(3-naphthalen-2-yl-1~{H}-1,2,4-triazol-5-yl)oxane-3,4-diol, Glycogen phosphorylase, muscle form, ... | | Authors: | Solovou, T.G.A, Kyriakis, E, Stravodimos, G.A, Kantsadi, A.L, Chatzileontiadou, D.S, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Nanomolar Inhibitors of Glycogen Phosphorylase Based on beta-d-Glucosaminyl Heterocycles: A Combined Synthetic, Enzyme Kinetic, and Protein Crystallography Study.
J. Med. Chem., 60, 2017
|
|
5B8F
 
 | |
5OX4
 
 | | Glycogen Phosphorylase in complex with CK900 | | Descriptor: | (2~{S},3~{R},4~{R},5~{S},6~{R})-2-[5-(4-aminophenyl)-4~{H}-1,2,4-triazol-3-yl]-6-(hydroxymethyl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Kantsadi, A.L, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2017-09-05 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the beta-pocket of the active site of human liver glycogen phosphorylase with 3-(C-beta-d-glucopyranosyl)-5-(4-substituted-phenyl)-1, 2, 4-triazole inhibitors.
Bioorg. Chem., 77, 2018
|
|
6SEA
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in deep mode | | Descriptor: | ACETATE ION, Beta-galactosidase, SODIUM ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
5OCE
 
 | | THE MOLECULAR MECHANISM OF SUBSTRATE RECOGNITION AND CATALYSIS OF THE MEMBRANE ACYLTRANSFERASE PatA -- Complex of PatA with palmitate, mannose, and palmitoyl-6-mannose | | Descriptor: | PALMITIC ACID, Phosphatidylinositol mannoside acyltransferase, [(2~{R},3~{S},4~{S},5~{S},6~{S})-3,4,5,6-tetrakis(oxidanyl)oxan-2-yl]methyl hexadecanoate, ... | | Authors: | Albesa-Jove, D, Tersa, M, Guerin, M.E. | | Deposit date: | 2017-06-30 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Molecular Mechanism of Substrate Recognition and Catalysis of the Membrane Acyltransferase PatA from Mycobacteria.
ACS Chem. Biol., 13, 2018
|
|
5I4F
 
 | | scFv 2D10 complexed with alpha 1,6 mannobiose | | Descriptor: | alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose, scFv 2D10 | | Authors: | Vashisht, S, Kumar, A, Kaur, K.J, Salunke, D.M. | | Deposit date: | 2016-02-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Antibodies Can Exploit Molecular Crowding to Bind New Antigens at Noncanonical Paratope Positions
CHEMISTRYSELECT, 1, 2016
|
|
