3G9C
 
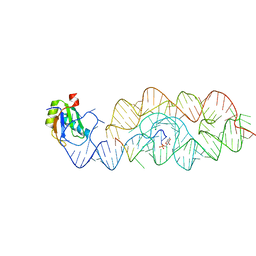 | | Crystal structure of the product Bacillus anthracis glmS ribozyme | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, GLMS RIBOZYME, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Cochrane, J.C, Lipchock, S.V, Smith, K.D. | | Deposit date: | 2009-02-13 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme
Biochemistry, 48, 2009
|
|
3G2Y
 
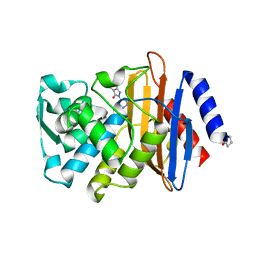 | | CTX-M-9 class A beta-lactamase complexed with compound 1 (GF4) | | Descriptor: | 4-ethyl-5-methyl-2-(1H-tetrazol-5-yl)-1,2-dihydro-3H-pyrazol-3-one, Beta-lactamase CTX-M-9a, DIMETHYL SULFOXIDE | | Authors: | Chen, Y, Shoichet, B.K. | | Deposit date: | 2009-02-01 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Molecular docking and ligand specificity in fragment-based inhibitor discovery
Nat.Chem.Biol., 5, 2009
|
|
3G50
 
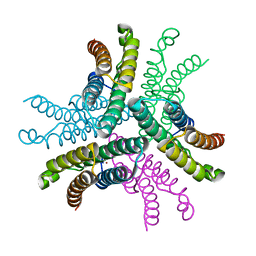 | | Crystal Structure of NiSOD D3A mutant at 1.9 A | | Descriptor: | NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Garman, S.C, Guce, A.I, Herbst, R.W, Bryngelson, P.A, Cabelli, D.E, Higgins, K.A, Ryan, K.C, Maroney, M.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of conserved tyrosine residues in NiSOD catalysis: a case of convergent evolution
Biochemistry, 48, 2009
|
|
3G5B
 
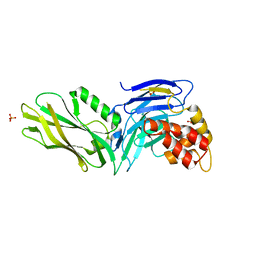 | | The structure of UNC5b cytoplasmic domain | | Descriptor: | Netrin receptor UNC5B, PHOSPHATE ION | | Authors: | Wang, R, Wei, Z, Zhang, M. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Autoinhibition of UNC5b revealed by the cytoplasmic domain structure of the receptor
Mol.Cell, 33, 2009
|
|
3GBA
 
 | | X-ray structure of iGluR5 ligand-binding core (S1S2) in complex with dysiherbaine at 1.35A resolution | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Frydenvang, K, Naur, P, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Full Domain Closure of the Ligand-binding Core of the Ionotropic Glutamate Receptor iGluR5 Induced by the High Affinity Agonist Dysiherbaine and the Functional Antagonist 8,9-Dideoxyneodysiherbaine
J.Biol.Chem., 284, 2009
|
|
3GDI
 
 | |
3GDN
 
 | | Almond hydroxynitrile lyase in complex with benzaldehyde | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dreveny, I, Gruber, K, Kratky, C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Substrate binding in the FAD-dependent hydroxynitrile lyase from almond provides insight into the mechanism of cyanohydrin formation and explains the absence of dehydrogenation activity.
Biochemistry, 48, 2009
|
|
3G5T
 
 | | Crystal structure of trans-aconitate 3-methyltransferase from yeast | | Descriptor: | (2E)-2-(2-methoxy-2-oxoethyl)but-2-enedioic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Crystal structure of trans-aconitate 3-methyltransferase from yeast
To be Published
|
|
3GF7
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum apoprotein | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, SULFATE ION | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GHE
 
 | |
3FWG
 
 | | Ferric camphor bound Cytochrome P450cam, Arg365Leu, Glu366Gln, monoclinic crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Schlichting, I, Von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FRR
 
 | |
3FRW
 
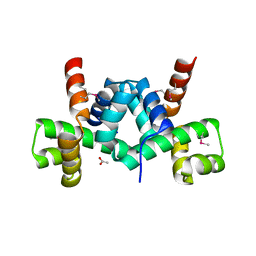 | | Crystal structure of putative TrpR protein from Ruminococcus obeum | | Descriptor: | ACETATE ION, Putative Trp repressor protein | | Authors: | Osipiuk, J, Keigher, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-08 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of putative TrpR protein from Ruminococcus obeum.
To be Published
|
|
3FSE
 
 | |
3FQN
 
 | |
3FQC
 
 | | Staphylococcus aureus dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-[3-(3,4,5-trimethoxyphenyl)pent-1-ynyl]-6-methylpyrimidine (UCP115A) | | Descriptor: | 6-methyl-5-[3-methyl-3-(3,4,5-trimethoxyphenyl)but-1-yn-1-yl]pyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2009-01-07 | | Release date: | 2009-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance.
J.Mol.Biol., 387, 2009
|
|
3FR1
 
 | |
3FUT
 
 | | Apo-form of T. thermophilus 16S rRNA A1518 and A1519 methyltransferase (KsgA) in space group P21212 | | Descriptor: | Dimethyladenosine transferase | | Authors: | Demirci, H, Belardinelli, R, Seri, E, Gregory, S.T, Gualerzi, C, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2009-01-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural rearrangements in the active site of the Thermus thermophilus 16S rRNA methyltransferase KsgA in a binary complex with 5'-methylthioadenosine.
J.Mol.Biol., 388, 2009
|
|
3FVE
 
 | | Crystal structure of diaminopimelate epimerase Mycobacterium tuberculosis DapF | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Diaminopimelate epimerase, GLYCEROL | | Authors: | Usha, V, Dover, L.G, Roper, D.I, Futterer, K, Besra, G.S. | | Deposit date: | 2009-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the diaminopimelate epimerase DapF from Mycobacterium tuberculosis
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3FVB
 
 | |
3FWF
 
 | | Ferric camphor bound cytochrome P450cam containing a Selenocysteine as the 5th heme ligand, monoclinic crystal form | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Schlichting, I, Von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FXK
 
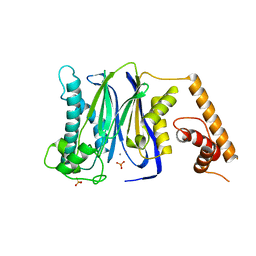 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 10 mM of Mn2+ | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FWU
 
 | | Crystal structure of Leishmania major MIF1 | | Descriptor: | Macrophage migration inhibitory factor-like protein | | Authors: | RIchardson, J.M, Walkinshaw, M.D. | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Leishmania major orthologues of macrophage migration inhibitory factor
Biochem.Biophys.Res.Commun., 380, 2009
|
|
3FX3
 
 | | Structure of a putative cAMP-binding regulatory protein from Silicibacter pomeroyi DSS-3 | | Descriptor: | Cyclic nucleotide-binding protein, GLYCEROL, PHOSPHATE ION | | Authors: | Cuff, M.E, Zhou, M, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a putative cAMP-binding regulatory protein from Silicibacter pomeroyi DSS-3
TO BE PUBLISHED
|
|
3FXJ
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Phosphate at 3 mM of Mn2+ | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
