6H2L
 
 | |
5MYJ
 
 | | Structure of 70S ribosome from Lactococcus lactis | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Franken, L.E, Oostergetel, G.T, Pijning, T, Puri, P, Boekema, E.J, Poolman, B, Guskov, A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy.
Nat Commun, 8, 2017
|
|
5U8K
 
 | | RitR mutant - C128S | | Descriptor: | Response regulator | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2016-12-14 | | Release date: | 2017-12-20 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
5MBA
 
 | | BINDING MODE OF AZIDE TO FERRIC APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | AZIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1991-01-14 | | Release date: | 1992-07-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding mode of azide to ferric Aplysia limacina myoglobin. Crystallographic analysis at 1.9 A resolution.
J.Mol.Recog., 4, 1991
|
|
3OYZ
 
 | | Haloferax volcanii Malate Synthase Pyruvate/Acetyl-CoA Ternary Complex | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C, Neighbor, A, Thomas, G, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-09-24 | | Release date: | 2011-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
1DUS
 
 | | MJ0882-A hypothetical protein from M. jannaschii | | Descriptor: | MJ0882 | | Authors: | Hung, L, Huang, L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based experimental confirmation of biochemical function to a methyltransferase, MJ0882, from hyperthermophile Methanococcus jannaschii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
6SWD
 
 | | IC2 body model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
6UOM
 
 | | Y271G DNA polymerase beta ternary complex with templating adenine and incoming r8-oxo-GTP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXYCYTIDINE-5'-MONOPHOSPHATE, 8-OXO-GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Smith, M.R, Freudenthal, B.D. | | Deposit date: | 2019-10-15 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular and structural characterization of oxidized ribonucleotide insertion into DNA by human DNA polymerase beta.
J.Biol.Chem., 295, 2020
|
|
6SKI
 
 | |
6IPU
 
 | |
6SJL
 
 | |
1HE1
 
 | | Crystal structure of the complex between the GAP domain of the Pseudomonas aeruginosa ExoS toxin and human Rac | | Descriptor: | ALUMINUM FLUORIDE, EXOENZYME S, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wurtele, M, Wolf, E, Pederson, K.J, Buchwald, G, Ahmadian, M.R, Barbieri, J.T, Wittinghofer, A. | | Deposit date: | 2000-11-18 | | Release date: | 2001-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How the Pseudomonas Aeruginosa Exos Toxin Downregulates Rac
Nat.Struct.Biol., 8, 2001
|
|
2K0J
 
 | | Solution structure of CaM complexed to DRP1p | | Descriptor: | CALCIUM ION, LANTHANUM (III) ION, calmodulin | | Authors: | Bertini, I, Luchinat, C, Parigi, G, Yuan, J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2008-02-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Accurate solution structures of proteins from X-ray data and a minimal set of NMR data: calmodulin-peptide complexes as examples.
J.Am.Chem.Soc., 131, 2009
|
|
6I4T
 
 | | Crystal structure of the disease-causing I445M mutant of the human dihydrolipoamide dehydrogenase | | Descriptor: | Dihydrolipoyl dehydrogenase, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Szabo, E, Wilk, P, Zambo, Z, Torocsik, B, Weiss, M.S, Adam-Vizi, V, Ambrus, A. | | Deposit date: | 2018-11-10 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Underlying molecular alterations in human dihydrolipoamide dehydrogenase deficiency revealed by structural analyses of disease-causing enzyme variants.
Hum.Mol.Genet., 28, 2019
|
|
6R9M
 
 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AAGGAA RNA | | Descriptor: | AAGGAA RNA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.329 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6R9Q
 
 | | Structure of Saccharomyces cerevisiae apo Pan2 pseudoubiquitin hydrolase-RNA exonuclease (UCH-Exo) module in complex with AACCAA RNA | | Descriptor: | AACCAA, PAN2-PAN3 deadenylation complex catalytic subunit PAN2 | | Authors: | Tang, T.T.L, Stowell, J.A.W, Hill, C.H, Passmore, L.A. | | Deposit date: | 2019-04-03 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.079 Å) | | Cite: | The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6V35
 
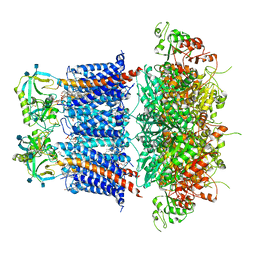 | | Cryo-EM structure of Ca2+-free hsSlo1-beta4 channel complex | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
6V22
 
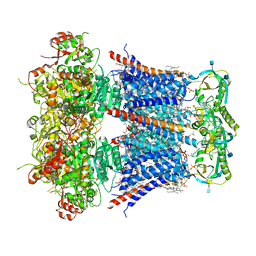 | | Cryo-EM structure of Ca2+-bound hsSlo1-beta4 channel complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tao, X, MacKinnon, R. | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular structures of the human Slo1 K + channel in complex with beta 4.
Elife, 8, 2019
|
|
6H55
 
 | | core of the human pyruvate dehydrogenase (E2) | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
5NN8
 
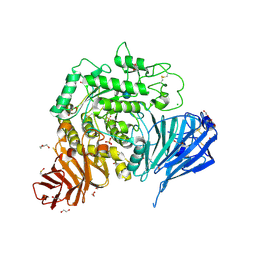 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with acarbose | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
5MJ5
 
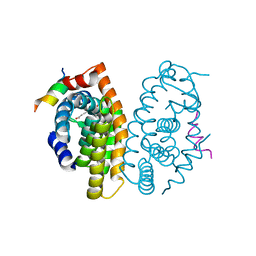 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetichonokiol derivative 3 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-[3-(phenylmethyl)phenyl]phenyl]prop-2-enoic acid, LYS-HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
2BNH
 
 | | PORCINE RIBONUCLEASE INHIBITOR | | Descriptor: | RIBONUCLEASE INHIBITOR | | Authors: | Kobe, B, Deisenhofer, J. | | Deposit date: | 1996-06-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A.
J.Mol.Biol., 264, 1996
|
|
1DRF
 
 | |
3K24
 
 | | Crystal structure of mature apo-Cathepsin L C25A mutant in complex with Gln-Leu-Ala peptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-6)-beta-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin L1, ... | | Authors: | Adams-Cioaba, M.A, Krupa, J.C, Mort, J.S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2010-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition and cleavage of histone H3 by cathepsin L.
Nat Commun, 2, 2011
|
|
