7AJ8
 
 | | Structure of DYRK1A in complex with compound 25 | | Descriptor: | 4-(1-benzofuran-5-yl)pyridin-2-amine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AJ7
 
 | | Structure of DYRK1A in complex with compound 16 | | Descriptor: | 4-(3-methylbenzimidazol-5-yl)pyridine-2,6-diamine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AJS
 
 | | Structure of DYRK1A in complex with compound 33 | | Descriptor: | 4-(2-methyl-1-benzofuran-5-yl)pyridine-2,6-diamine, DIMETHYL SULFOXIDE, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ... | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
4R5X
 
 | |
7AJ5
 
 | | Structure of DYRK1A in complex with compound 10 | | Descriptor: | 4-(1-benzofuran-5-yl)pyridine-2,6-diamine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
7AJM
 
 | | Structure of DYRK1A in complex with compound 32 | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}-[6-azanyl-4-(1-benzofuran-5-yl)pyridin-2-yl]-2-(methylamino)ethanamide | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
3CHV
 
 | |
4R1M
 
 | |
7AJA
 
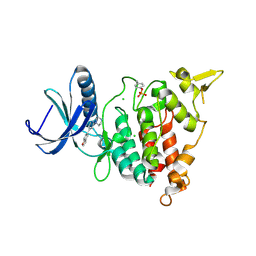 | | Structure of DYRK1A in complex with compound 28 | | Descriptor: | 1-[4-(1-benzofuran-5-yl)pyridin-2-yl]piperazine, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Dokurno, P, Surgenor, A.E, Kotschy, A. | | Deposit date: | 2020-09-28 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective DYRK1A Inhibitors.
J.Med.Chem., 64, 2021
|
|
4R8O
 
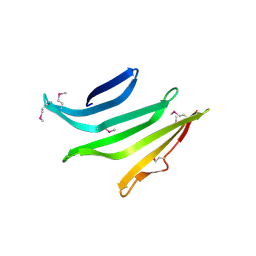 | |
3CGG
 
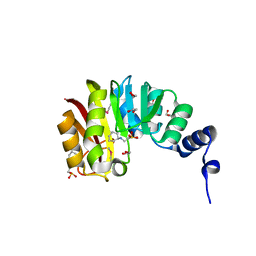 | |
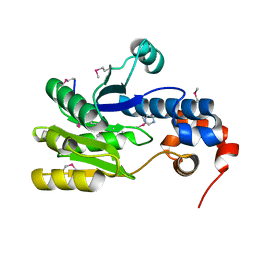
3CGX
 
 | |
3CSW
 
 | |
3CWR
 
 | |
4R1S
 
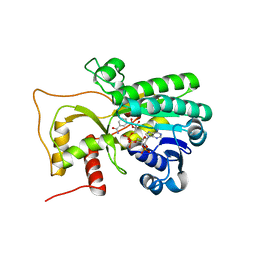 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, cinnamoyl CoA reductase | | Authors: | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
3D02
 
 | |
3D2E
 
 | | Crystal structure of a complex of Sse1p and Hsp70, Selenomethionine-labeled crystals | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Heat shock 70 kDa protein 1, ... | | Authors: | Polier, S, Bracher, A. | | Deposit date: | 2008-05-08 | | Release date: | 2008-06-17 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding.
Cell(Cambridge,Mass.), 133, 2008
|
|
7B2H
 
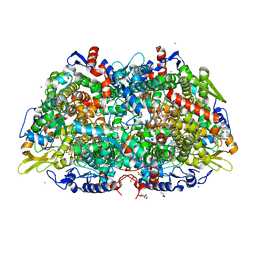 | | Crystal structure of the methyl-coenzyme M reductase from Methanothermobacter Marburgensis derivatized with xenon | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-27 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
3CEB
 
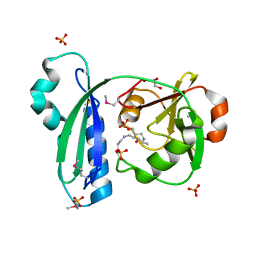 | |
3D4O
 
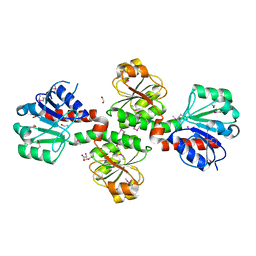 | |
3CF9
 
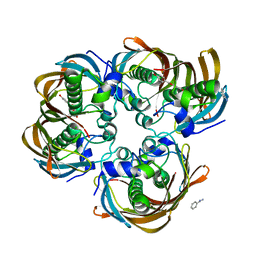 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with apigenin | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, BENZAMIDINE, ... | | Authors: | Zhang, L, Wu, D, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2008-03-03 | | Release date: | 2008-12-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three flavonoids targeting the beta-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay
Protein Sci., 17, 2008
|
|
3CEC
 
 | |
7B5W
 
 | |
3CU2
 
 | |
3D00
 
 | |
