4F65
 
 | | Crystal structure of Human Fibroblast Growth Factor Receptor 1 Kinase domain in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N~2~-[(3-methyl-1,2-oxazol-5-yl)methyl]-N~4~-[3-(2-phenylethyl)-1H-pyrazol-5-yl]pyrimidine-2,4-diamine, Fibroblast growth factor receptor 1, ... | | Authors: | Norman, R.A, Breed, J, Ogg, D. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Protein-Ligand Crystal Structures Can Guide the Design of Selective Inhibitors of the FGFR Tyrosine Kinase.
J.Med.Chem., 55, 2012
|
|
4F63
 
 | | Crystal structure of Human Fibroblast Growth Factor Receptor 1 Kinase domain in complex with compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N~4~-(3-methyl-1H-pyrazol-5-yl)-N~2~-[2-(pyridin-3-yl)ethyl]pyrimidine-2,4-diamine, Fibroblast growth factor receptor 1 | | Authors: | Norman, R.A, Breed, J, Ogg, D. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Protein-Ligand Crystal Structures Can Guide the Design of Selective Inhibitors of the FGFR Tyrosine Kinase.
J.Med.Chem., 55, 2012
|
|
4F64
 
 | | Crystal structure of Human Fibroblast Growth Factor Receptor 1 Kinase domain in complex with compound 6 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N~4~-[3-(3-methoxypropyl)-1H-pyrazol-5-yl]-N~2~-[(3-methyl-1,2-oxazol-5-yl)methyl]pyrimidine-2,4-diamine, Fibroblast growth factor receptor 1, ... | | Authors: | Norman, R.A, Breed, J, Ogg, D. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Protein-Ligand Crystal Structures Can Guide the Design of Selective Inhibitors of the FGFR Tyrosine Kinase.
J.Med.Chem., 55, 2012
|
|
5ZHG
 
 | |
8PF9
 
 | | Galectin-3C in complex with a triazolesulfane derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[oxidanyl(phenyl)-$l^{3}-sulfanyl]-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8PBF
 
 | | Galectin-3C in complex with a triazolesulfane derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-(4-phenylsulfanyl-1,2,3-triazol-1-yl)oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, THIOCYANATE ION | | Authors: | Kumar, R, Mahanti, M. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
8PFF
 
 | | Galectin-3C in complex with a triazolesulfone derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-(phenylsulfonyl)-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
7NSB
 
 | | Supramolecular assembly module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | Descriptor: | Glucose-induced degradation protein 7, Glucose-induced degradation protein 8, Vacuolar import and degradation protein 30 | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
1MVC
 
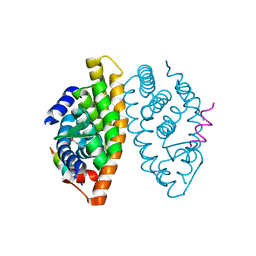 | | Crystal structure of the human RXR alpha ligand binding domain bound to the synthetic agonist compound BMS 649 and a coactivator peptide | | Descriptor: | 4-[2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-[1,3]DIOXOLAN-2-YL]-BENZOIC ACID, Nuclear receptor coactivator 2, RXR retinoid X receptor | | Authors: | Egea, P.F, Mitschler, A, Moras, D. | | Deposit date: | 2002-09-24 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Recognition of Agonist Ligands by RXRs
MOL.ENDOCRINOL., 16, 2002
|
|
5M3A
 
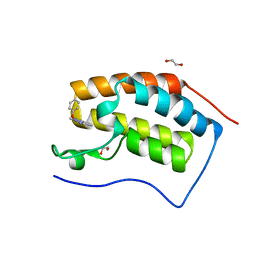 | | Crystal structure of BRD4 BROMODOMAIN 1 IN COMPLEX WITH LIGAND 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-6-(1-methyl-5-phenoxy-pyrazol-4-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4 | | Authors: | Kessler, D, Mayer, M, Engelhardt, H, Wolkerstorfer, B, Geist, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Direct NMR Probing of Hydration Shells of Protein Ligand Interfaces and Its Application to Drug Design.
J. Med. Chem., 60, 2017
|
|
1NQ7
 
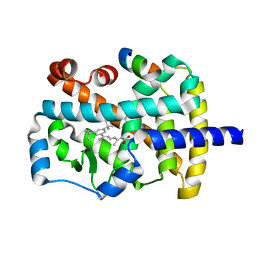 | | Characterization of ligands for the orphan nuclear receptor RORbeta | | Descriptor: | 7-(3,5-DITERT-BUTYLPHENYL)-3-METHYLOCTA-2,4,6-TRIENOIC ACID, NUCLEAR RECEPTOR ROR-BETA, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Stehlin-Gaon, C, Willmann, D, Sanglier, S, Van Dorsselaer, A, Renaud, J.-P, Moras, D, Schuele, R. | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | All-trans retinoic acid is a ligand for the orphan nuclear receptor RORbeta
Nat.Struct.Biol., 10, 2003
|
|
6VOE
 
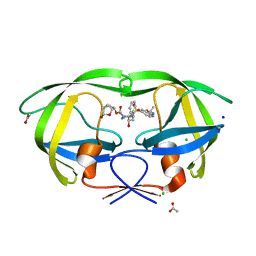 | | HIV-1 wild type protease with GRL-019-17A, a tricyclic cyclohexane fused tetrahydrofuranofuran (CHf-THF) derivative as the P2 ligand and a aminobenzothiazole(Abt)-based P2'-ligand | | Descriptor: | (1S,3aR,5S,6R,7aS)-octahydro-1,6-epoxy-2-benzofuran-5-yl {(2S,3R)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]-1-phenylbutan-2-yl}carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2020-01-30 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Design of Highly Potent HIV-1 Protease Inhibitors Containing New Tricyclic Ring P2-Ligands: Design, Synthesis, Biological, and X-ray Structural Studies.
J.Med.Chem., 63, 2020
|
|
8F3C
 
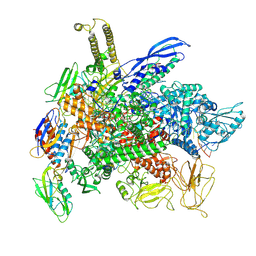 | | Cryo-EM consensus structure of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase minus preQ1 ligand | | Descriptor: | DNA (38-MER), DNA (39-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1MV9
 
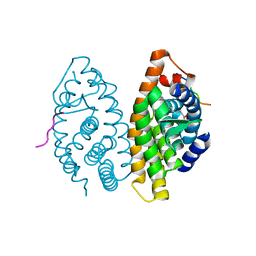 | | Crystal Structure of the human RXR alpha ligand binding domain bound to the eicosanoid DHA (Docosa Hexaenoic Acid) and a coactivator peptide | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Nuclear receptor coactivator 2, RXR retinoid X receptor | | Authors: | Egea, P.F, Mitschler, A, Moras, D. | | Deposit date: | 2002-09-24 | | Release date: | 2002-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Recognition of Agonist Ligands by RXRs
MOL.ENDOCRINOL., 16, 2002
|
|
2OH9
 
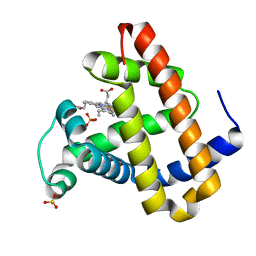 | | Myoglobin cavity mutant V68W | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Phillips Jr, G.N, Li, T, Brucker, E.A, Soman, J, Olson, J.S. | | Deposit date: | 2007-01-09 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand pathways in myoglobin: A review of trp cavity mutations.
Iubmb Life, 59, 2007
|
|
6CZD
 
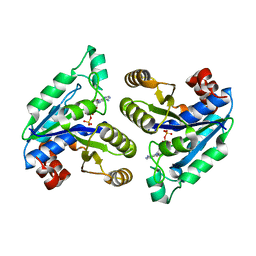 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent dethiobiotin synthetase BioD, MAGNESIUM ION | | Authors: | Thompson, A.P, Wegener, K.L, Bruning, J.B, Polyak, S.W. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Precipitant-ligand exchange technique reveals the ADP binding mode in Mycobacterium tuberculosis dethiobiotin synthetase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1U88
 
 | |
1U87
 
 | |
8BTT
 
 | | Structure of human RTCB | | Descriptor: | MANGANESE (II) ION, RNA-splicing ligase RtcB homolog | | Authors: | Kopp, J, Gerber, J.L, Peschek, J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic insights into activation of the human RNA ligase RTCB by Archease.
Nat Commun, 15, 2024
|
|
6KB0
 
 | | X-ray structure of human PPARalpha ligand binding domain-5,8,11,14-eicosatetraynoic acid (ETYA) co-crystals obtained by soaking | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, icosa-5,8,11,14-tetraynoic acid | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB2
 
 | | X-ray structure of human PPARalpha ligand binding domain-Wy14643 co-crystals obtained by soaking | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB9
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by cross-seeding | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB1
 
 | | X-ray structure of human PPARalpha ligand binding domain-tetradecylthioacetic acid (TTA) co-crystals obtained by soaking | | Descriptor: | 2-tetradecylsulfanylethanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KAZ
 
 | | X-ray structure of human PPARalpha ligand binding domain-pemafibrate co-crystals obtained by soaking | | Descriptor: | (2~{R})-2-[3-[[1,3-benzoxazol-2-yl-[3-(4-methoxyphenoxy)propyl]amino]methyl]phenoxy]butanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6KB5
 
 | | X-ray structure of human PPARalpha ligand binding domain-5,8,11,14-eicosatetraynoic Acid (ETYA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, Peroxisome proliferator-activated receptor alpha, icosa-5,8,11,14-tetraynoic acid | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
