7RYA
 
 | | S. CEREVISIAE CYP51 I471T MUTANT COMPLEXED WITH ITRACONAZOLE | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
7RY9
 
 | | S. CEREVISIAE CYP51 I471T mutant COMPLEXED WITH Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, TETRAETHYLENE GLYCOL, ... | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
7RY8
 
 | | S. CEREVISIAE CYP51 Y140H mutant COMPLEXED WITH Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PENTAETHYLENE GLYCOL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
7RYB
 
 | | S. CEREVISIAE CYP51 Y140H/I471T - double mutant COMPLEXED WITH Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
7ZBO
 
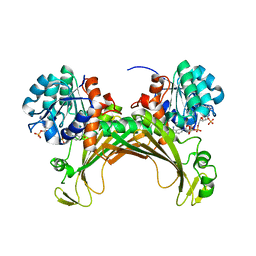 | | Amine Dehydrogenase MATOUAmDH2 in complex with NADP+ | | Descriptor: | Amine Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bennett, M, Ducrot, L, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure and Mutation of the Native Amine Dehydrogenase MATOUAmDH2.
Chembiochem, 23, 2022
|
|
2MQ8
 
 | | Solution NMR Structure of De novo designed protein LFR1 1 with ferredoxin fold, Northeast Structural Genomics Consortium (NESG) Target OR414 | | Descriptor: | De novo designed protein LFR1 | | Authors: | Liu, G, Lin, Y, Koga, N, Koga, R, Xiao, R, Janjua, H, Pederson, K, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-06-12 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Control over overall shape and size in de novo designed proteins.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6EON
 
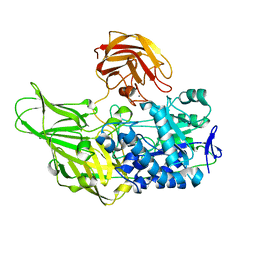 | | Galactanase BT0290 | | Descriptor: | Beta-galactosidase, CALCIUM ION, alpha-D-galactopyranose | | Authors: | Basle, A, Munoz, J, Gilbert, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6XEH
 
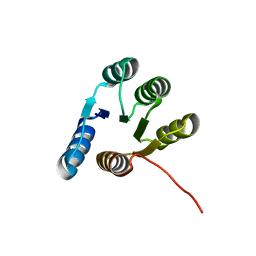 | |
6EUI
 
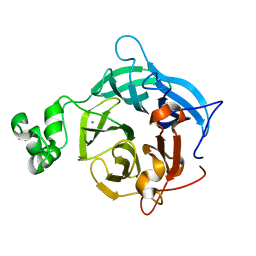 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactose | | Descriptor: | Beta-glucanase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUJ
 
 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUF
 
 | | The GH43, Beta 1,3 Galactosidase, BT0265 | | Descriptor: | Beta-glucanase, alpha-L-arabinofuranose-(1-3)-[alpha-L-arabinofuranose-(1-4)][beta-D-glucopyranuronic acid-(1-6)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose, alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-6)-[alpha-L-arabinofuranose-(1-3)][alpha-L-arabinofuranose-(1-4)]beta-D-galactopyranose-(1-6)-beta-D-galactopyranose | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUH
 
 | | The GH43, Beta 1,3 Galactosidase, BT3683 with galactodeoxynojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, Beta-glucanase, CALCIUM ION | | Authors: | Cartmell, A, Gilbert, H.J. | | Deposit date: | 2017-10-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
6EUG
 
 | |
5T47
 
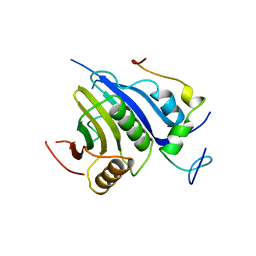 | | Crystal structure of the D. melanogaster eIF4E-eIF4G complex | | Descriptor: | Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4G, isoform A, ... | | Authors: | Gruener, S, Peter, D, Weber, R, Wohlbold, L, Chung, M.-Y, Weichenrieder, O, Valkov, E, Igreja, C, Izaurralde, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Mol.Cell, 64, 2016
|
|
5T48
 
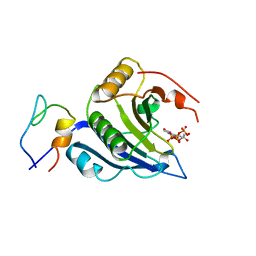 | | Crystal structure of the D. melanogaster eIF4E-eIF4G complex without lateral contact | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4G, ... | | Authors: | Gruener, S, Peter, D, Weber, R, Wohlbold, L, Chung, M.-Y, Weichenrieder, O, Valkov, E, Igreja, C, Izaurralde, E. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structures of eIF4E-eIF4G Complexes Reveal an Extended Interface to Regulate Translation Initiation.
Mol.Cell, 64, 2016
|
|
8AV5
 
 | |
7ZNM
 
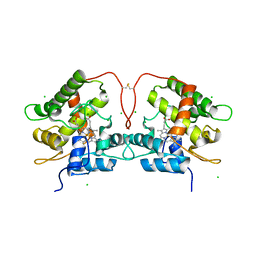 | | Artificial Unspecific Peroxygenase expressed in Pichia pastoris at 2.01 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Artificial Unspecific Peroxygenase, ... | | Authors: | Robinson, W.X.Q, Mielke, T, Grogan, G. | | Deposit date: | 2022-04-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Comparing the Catalytic and Structural Characteristics of a 'Short' Unspecific Peroxygenase (UPO) Expressed in Pichia pastoris and Escherichia coli.
Chembiochem, 24, 2023
|
|
7ZNV
 
 | | Artificial Unspecific Peroxygenase expressed in Pichia pastoris at 1.21 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, ... | | Authors: | Robinson, W.X.Q, Mielke, T, Grogan, G. | | Deposit date: | 2022-04-22 | | Release date: | 2023-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Comparing the Catalytic and Structural Characteristics of a 'Short' Unspecific Peroxygenase (UPO) Expressed in Pichia pastoris and Escherichia coli.
Chembiochem, 24, 2023
|
|
7ZNW
 
 | |
8A3X
 
 | | Imine Reductase from Ensifer adhaerens in complex with NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NAD_binding_2 domain-containing protein, POTASSIUM ION | | Authors: | Gilio, A.K, Grogan, G. | | Deposit date: | 2022-06-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Reductive Aminase Switches to Imine Reductase Mode for a Bulky Amine Substrate.
Acs Catalysis, 13, 2023
|
|
8A5Z
 
 | |
1GV5
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 | | Descriptor: | MYB PROTO-ONCOGENE PROTEIN, SODIUM ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-06 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
6ZBI
 
 | | Ternary complex of Calmodulin bound to 2 molecules of NHE1 | | Descriptor: | CALCIUM ION, Calmodulin-1, Sodium/hydrogen exchanger 1 | | Authors: | Prestel, A, Kragelund, B.B, Pedersen, E.S, Pedersen, S.F, Sjoegaard-Frich, L.M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic Na + /H + exchanger 1 (NHE1) - calmodulin complexes of varying stoichiometry and structure regulate Ca 2+ -dependent NHE1 activation.
Elife, 10, 2021
|
|
1GVD
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 V103L MUTANT | | Descriptor: | AMMONIUM ION, MYB PROTO-ONCOGENE PROTEIN, SULFATE ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
8QHC
 
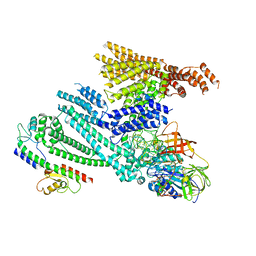 | | Cryo-EM structure of SidH from Legionella pneumophila in complex with LubX | | Descriptor: | E3 ubiquitin--protein ligase, Elongation factor Tu, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sharma, R, Adams, M, Bhogaraju, S. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the toxicity of Legionella pneumophila effector SidH.
Nat Commun, 14, 2023
|
|
