3HYX
 
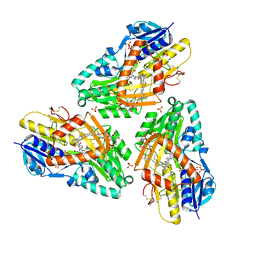 | | 3-D X-Ray structure of the sulfide:quinone oxidoreductase from Aquifex aeolicus in complex with Aurachin C | | Descriptor: | 1-hydroxy-2-methyl-3-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]quinolin-4(1H)-one, DODECYL-BETA-D-MALTOSIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Marcia, M, Ermler, U, Peng, G.H, Michel, H. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1T0M
 
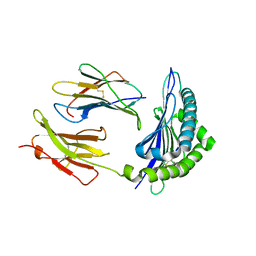 | | Conformational switch in polymorphic H-2K molecules containing an HSV peptide | | Descriptor: | Beta-2-microglobulin, Glycoprotein B, H-2 class I histocompatibility antigen, ... | | Authors: | Webb, A.I, Borg, N.A, Dunstone, M.A, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Carbone, F.R, Bottomley, S.P, Purcell, A.W, Rossjohn, J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of H-2K(b) and K(bm8) complexed to a herpes simplex virus determinant: evidence for a conformational switch that governs T cell repertoire selection and viral resistance.
J Immunol., 173, 2004
|
|
3H65
 
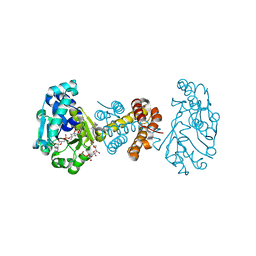 | | The Crystal Structure of C176A Mutated [Fe]-Hydrogenase (Hmd) Holoenzyme in Complex with Methylenetetrahydromethanopterin | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, ... | | Authors: | Hiromoto, T, Warkentin, E, Shima, S, Ermler, U. | | Deposit date: | 2009-04-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of an [Fe]-hydrogenase-substrate complex reveals the framework for H2 activation.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3IQF
 
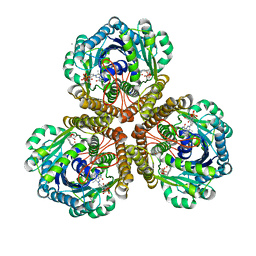 | | Structure of F420 dependent methylene-tetrahydromethanopterin dehydrogenase in complex with methenyl-tetrahydromethanopterin | | Descriptor: | 1-{4-[(6S,6aR,7R)-3-amino-6,7-dimethyl-1-oxo-1,2,5,6,6a,7-hexahydro-8H-imidazo[1,5-f]pteridin-10-ium-8-yl]phenyl}-1-deoxy-5-O-{5-O-[(S)-{[(1S)-1,3-dicarboxypropyl]oxy}(hydroxy)phosphoryl]-alpha-D-ribofuranosyl}-D-ribitol, CALCIUM ION, F420-dependent methylenetetrahydromethanopterin dehydrogenase, ... | | Authors: | Ceh, K.E, Demmer, U, Warkentin, E, Moll, J, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2009-08-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the hydride transfer mechanism in F(420)-dependent methylenetetrahydromethanopterin dehydrogenase
Biochemistry, 48, 2009
|
|
1WZ9
 
 | | The 2.1 A structure of a tumour suppressing serpin | | Descriptor: | Maspin precursor, SULFATE ION | | Authors: | Law, R.H, Irving, J.A, Buckle, A.M, Ruzyla, K, Buzza, M, Bashtannyk-Puhalovich, T.A, Beddoe, T.C, Kim, N, Worrall, D.M, Bottomley, S.P, Bird, P.I, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2005-03-03 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
3B4V
 
 | | X-Ray structure of Activin in complex with FSTL3 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Inhibin beta A chain, ... | | Authors: | Thompson, T.B. | | Deposit date: | 2007-10-24 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The structure of FSTL3.activin A complex. Differential binding of N-terminal domains influences follistatin-type antagonist specificity.
J.Biol.Chem., 283, 2008
|
|
1PCG
 
 | | Helix-stabilized cyclic peptides as selective inhibitors of steroid receptor-coactivator interactions | | Descriptor: | ESTRADIOL, estrogen receptor, peptide inhibitor | | Authors: | Leduc, A.M, Trent, J.O, Wittliff, J.L, Bramlett, K.S, Briggs, S.L, Chirgadze, N.Y, Wang, Y, Burris, T.P, Spatola, A.F. | | Deposit date: | 2003-05-16 | | Release date: | 2003-10-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Helix-stabilized cyclic peptides as selective inhibitors of steroid receptor-coactivator interactions
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3F47
 
 | | The Crystal Structure of [Fe]-Hydrogenase (Hmd) Holoenzyme from Methanocaldococcus jannaschii | | Descriptor: | 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, CARBON MONOXIDE, ... | | Authors: | Hiromoto, T, Pilak, O, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of C176A mutated [Fe]-hydrogenase suggests an acyl-iron ligation in the active site iron complex.
Febs Lett., 583, 2009
|
|
1QV9
 
 | | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: A methanogenic enzyme with an unusual quarternary structure | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Hagemeier, C.H, Shima, S, Thauer, R.K, Bourenkov, G, Bartunik, H.D, Ermler, U. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: a methanogenic enzyme with an unusual quarternary structure
J.Mol.Biol., 332, 2003
|
|
1OQJ
 
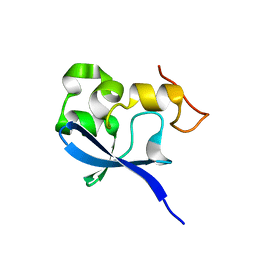 | | Crystal structure of the SAND domain from glucocorticoid modulatory element binding protein-1 (GMEB1) | | Descriptor: | Glucocorticoid Modulatory Element Binding protein-1, ZINC ION | | Authors: | Surdo, P.L, Bottomley, M.J, Sattler, M, Scheffzek, K. | | Deposit date: | 2003-03-10 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucocorticoid modulatory element binding protein-1 reveals deoxyribonucleic acid and zinc binding regions
MOL.ENDOCRINOL., 17, 2003
|
|
1RGV
 
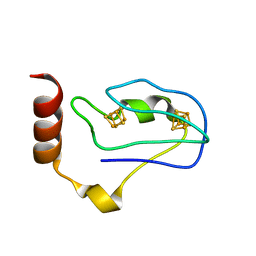 | | Crystal Structure of the Ferredoxin from Thauera aromatica | | Descriptor: | IRON/SULFUR CLUSTER, ferredoxin | | Authors: | Unciuleac, M, Boll, M, Warkentin, E, Ermler, U. | | Deposit date: | 2003-11-13 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallization of 4-hydroxybenzoyl-CoA reductase and the structure of its electron donor ferredoxin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RM6
 
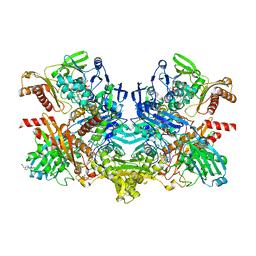 | | Structure of 4-hydroxybenzoyl-CoA reductase from Thauera aromatica | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxybenzoyl-CoA reductase alpha subunit, ... | | Authors: | Unciuleac, M, Warkentin, E, Page, C.C, Dutton, P.L, Boll, M, Ermler, U. | | Deposit date: | 2003-11-27 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Xanthine Oxidase-Related 4-Hydroxybenzoyl-CoA Reductase with an Additional [4Fe-4S] Cluster and an Inverted Electron Flow
Structure, 12, 2004
|
|
1RHC
 
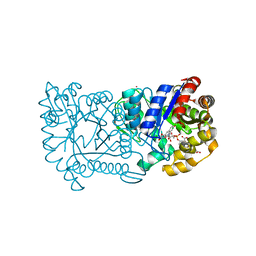 | | F420-dependent secondary alcohol dehydrogenase in complex with an F420-acetone adduct | | Descriptor: | ACETONE, CHLORIDE ION, COENZYME F420, ... | | Authors: | Aufhammer, S.W, Warkentin, E, Berk, H, Shima, S, Thauer, R.K, Ermler, U. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Coenzyme binding in f(420)-dependent secondary alcohol dehydrogenase, a member of the bacterial luciferase family.
Structure, 12, 2004
|
|
2V3A
 
 | | Crystal structure of rubredoxin reductase from Pseudomonas aeruginosa. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hagelueken, G, Wiehlmann, L, Adams, T.M, Kolmar, H, Heinz, D.W, Tuemmler, B, Schubert, W.-D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Electron Transfer Complex Rubredoxin - Rubredoxin Reductase from Pseudomonas Aeruginosa.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2V3B
 
 | | Crystal structure of the electron transfer complex rubredoxin - rubredoxin reductase from Pseudomonas aeruginosa. | | Descriptor: | FE (III) ION, FLAVIN-ADENINE DINUCLEOTIDE, RUBREDOXIN 2, ... | | Authors: | Hagelueken, G, Wiehlmann, L, Adams, T.M, Kolmar, H, Heinz, D.W, Tuemmler, B, Schubert, W.-D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Electron Transfer Complex Rubredoxin - Rubredoxin Reductase from Pseudomonas Aeruginosa.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2VH4
 
 | | Structure of a loop C-sheet serpin polymer | | Descriptor: | TENGPIN | | Authors: | Zhang, Q, Law, R.H.P, Bottomley, S.P, Whisstock, J.C, Buckle, A.M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Structural Basis for Loop C-Sheet Polymerization in Serpins.
J.Mol.Biol., 376, 2008
|
|
2Y37
 
 | | The discovery of novel, potent and highly selective inhibitors of inducible nitric oxide synthase (iNOS) | | Descriptor: | 2-[(1R)-3-amino-1-phenyl-propoxy]-4-chloro-benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Cheshire, D.R, Andrews, G, Beaton, H.G, Birkinshaw, T.N, Boughton-Smith, N, Connolly, S, Cook, T.R, Cooper, A, Cooper, S.L, Cox, D, Dixon, J, Gensmantel, N, Hamley, P.J, Harrison, R, Hartopp, P, Kack, H, Luker, T, Mete, A, Millichip, I, Nicholls, D.J, Pimm, A.D, St-Gallay, S.A, Wallace, A.V. | | Deposit date: | 2010-12-19 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Discovery of Novel, Potent and Highly Selective Inhibitors of Inducible Nitric Oxide Synthase (Inos).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3MM5
 
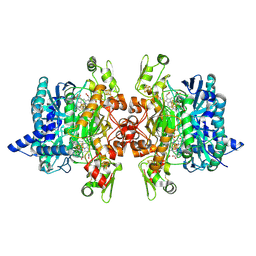 | | Dissimilatory sulfite reductase in complex with the substrate sulfite | | Descriptor: | IRON/SULFUR CLUSTER, SIROHEME, SULFITE ION, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3MPI
 
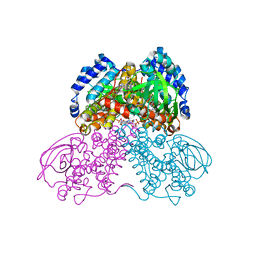 | | Structure of the glutaryl-coenzyme A dehydrogenase glutaryl-CoA complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, glutaryl-coenzyme A | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
3MK7
 
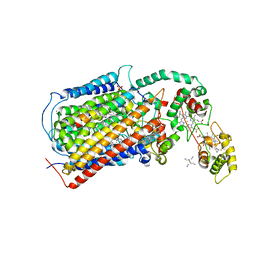 | | The structure of CBB3 cytochrome oxidase | | Descriptor: | 30-mer peptide, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Buschmann, S, Warkentin, E, Michel, H, Ermler, U. | | Deposit date: | 2010-04-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of cbb3 Cytochrome Oxidase Provides Insights into Proton Pumping
Science, 329, 2010
|
|
2MTQ
 
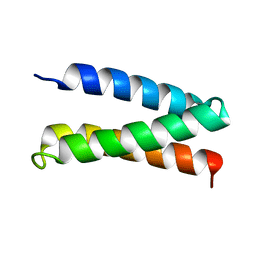 | | Solution Structure of a De Novo Designed Peptide that Sequesters Toxic Heavy Metals | | Descriptor: | Designed Peptide | | Authors: | Plegaria, J.S, Zuiderweg, E.R, Stemmler, T.L, Pecoraro, V.L. | | Deposit date: | 2014-08-28 | | Release date: | 2015-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Apoprotein Structure and Metal Binding Characterization of a de Novo Designed Peptide, alpha 3DIV, that Sequesters Toxic Heavy Metals.
Biochemistry, 54, 2015
|
|
3MM6
 
 | | Dissimilatory sulfite reductase cyanide complex | | Descriptor: | CYANIDE ION, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3OYX
 
 | | Haloferax volcanii Malate Synthase Magnesium/Glyoxylate Complex | | Descriptor: | CHLORIDE ION, GLYOXYLIC ACID, MAGNESIUM ION, ... | | Authors: | Howard, B.R, Bracken, C, Neighbor, A, Thomas, G, Lamlenn, K.K, Schubert, H.L, Whitby, F.G. | | Deposit date: | 2010-09-24 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of a halophilic archaeal malate synthase from Haloferax volcanii and comparisons with isoforms A and G.
Bmc Struct.Biol., 11, 2011
|
|
3MM9
 
 | | Dissimilatory sulfite reductase nitrite complex | | Descriptor: | IRON/SULFUR CLUSTER, NITRITE ION, SIROHEME, ... | | Authors: | Parey, K, Warkentin, E, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-07-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction cycle of the dissimilatory sulfite reductase from Archaeoglobus fulgidus.
Biochemistry, 49, 2010
|
|
3MPJ
 
 | | Structure of the glutaryl-coenzyme A dehydrogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, ... | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
