5NBK
 
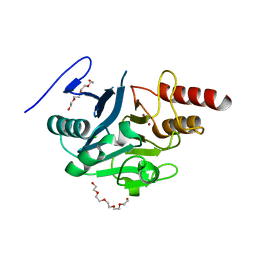 | | NDM-1 metallo-beta-lactamase: a parsimonious interpretation of the diffraction data | | Descriptor: | CHLORIDE ION, HEXAETHYLENE GLYCOL, Metallo-beta-lactamase type 2, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
5N20
 
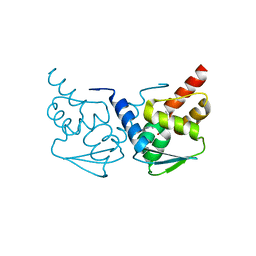 | | Crystal structure of the BCL6 BTB domain in complex with pyrazolo-pyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-2-[(3~{R})-3-(dimethylamino)pyrrolidin-1-yl]phenyl]ethanamide | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-02-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery of Pyrazolo[1,5-a]pyrimidine B-Cell Lymphoma 6 (BCL6) Binders and Optimization to High Affinity Macrocyclic Inhibitors.
J. Med. Chem., 60, 2017
|
|
5EU3
 
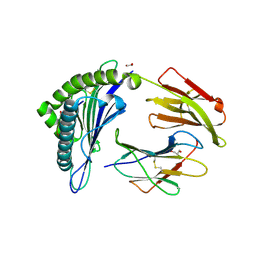 | | HLA Class I antigen | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Bianchi, V, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Molecular Switch Abrogates Glycoprotein 100 (gp100) T-cell Receptor (TCR) Targeting of a Human Melanoma Antigen.
J.Biol.Chem., 291, 2016
|
|
6IBP
 
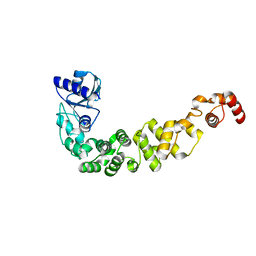 | | Structure of a psychrophilic CCA-adding enzyme at room temperature in ChipX microfluidic device | | Descriptor: | CCA-adding enzyme | | Authors: | de Wijn, R, Hennig, O, Rollet, K, Bluhm, A, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
197D
 
 | |
1A5Q
 
 | | P93A VARIANT OF BOVINE PANCREATIC RIBONUCLEASE A | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Pearson, M.A, Karplus, P.A, Dodge, R.W, Laity, J.H, Scheraga, H.A. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two mutants that have implications for the folding of bovine pancreatic ribonuclease A.
Protein Sci., 7, 1998
|
|
5I2V
 
 | |
5OUZ
 
 | | Metal free structure of Y40F SynFtn | | Descriptor: | Ferritin | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2017-08-25 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Reaction of O2with a diiron protein generates a mixed-valent Fe2+/Fe3+center and peroxide.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7UKP
 
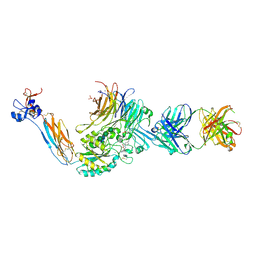 | | Integrin alpha IIB beta3 complex with a gantofiban analog | | Descriptor: | 10E5 Fab heavy chain, 10E5 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | Deposit date: | 2022-04-01 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.80112982 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
5EYC
 
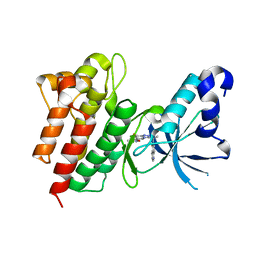 | | Crystal structure of c-Met in complex with naphthyridinone inhibitor 5 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(3-methyl-1,2-oxazol-5-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
6SEG
 
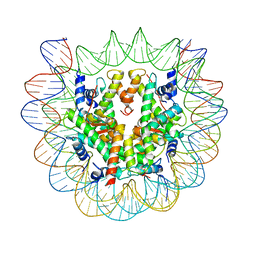 | | Class1: CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | DNA (145-MER), Histone H2A type 2-A, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
5ZZC
 
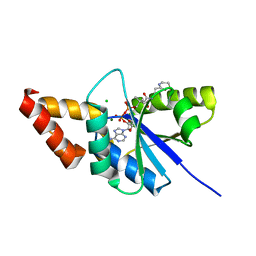 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 1.94A resolution | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Gupta, A, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-05-31 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 1.94 A resolution
To Be Published
|
|
6SE0
 
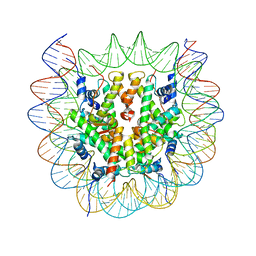 | | Class 1 : CENP-A nucleosome | | Descriptor: | DNA (145-MER), Histone H2A type 2-A, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
9GXA
 
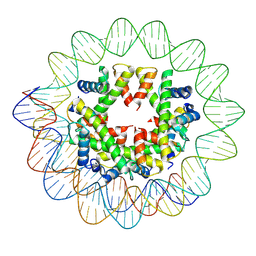 | | CENP-A/H4 di-tetrasome assembled on alpha-satellite DNA. | | Descriptor: | 147 bp alpha-satellite DNA, 147 bp human alpha-satellite DNA, Histone H3-like centromeric protein A, ... | | Authors: | Ali-Ahmad, A, Sekulic, N. | | Deposit date: | 2024-09-29 | | Release date: | 2025-01-29 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Non-nucleosomal (CENP-A/H4) 2 - DNA complexes as a possible platform for centromere organization.
Biorxiv, 2025
|
|
5OUW
 
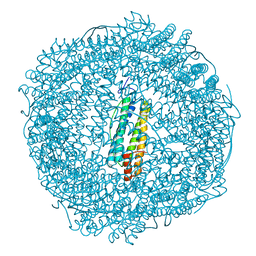 | | Metal free structure of SynFtn | | Descriptor: | Ferritin | | Authors: | Hemmings, A.M, Bradley, J.M. | | Deposit date: | 2017-08-25 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Reaction of O2with a diiron protein generates a mixed-valent Fe2+/Fe3+center and peroxide.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6A75
 
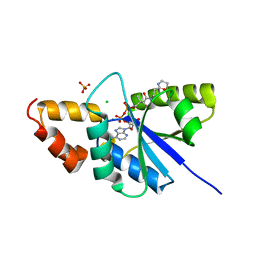 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.75 A resolution | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-07-02 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.75 A resolution
To Be Published
|
|
1AHL
 
 | | ANTHOPLEURIN-A,NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-A | | Authors: | Pallaghy, P.K, Scanlon, M.J, Monks, S.A, Norton, R.S. | | Deposit date: | 1994-10-28 | | Release date: | 1995-11-14 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure in solution of the polypeptide cardiac stimulant anthopleurin-A.
Biochemistry, 34, 1995
|
|
6SE6
 
 | | Class2 : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
5EYD
 
 | | Crystal structure of c-Met in complex with AMG 337 | | Descriptor: | 6-[(1~{R})-1-[8-fluoranyl-6-(1-methylpyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl]ethyl]-3-(2-methoxyethoxy)-1,6-naphthyridin-5-one, Hepatocyte growth factor receptor | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of (R)-6-(1-(8-Fluoro-6-(1-methyl-1H-pyrazol-4-yl)-[1,2,4]triazolo[4,3-a]pyridin-3-yl)ethyl)-3-(2-methoxyethoxy)-1,6-naphthyridin-5(6H)-one (AMG 337), a Potent and Selective Inhibitor of MET with High Unbound Target Coverage and Robust In Vivo Antitumor Activity.
J.Med.Chem., 59, 2016
|
|
6A7D
 
 | | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.74 A resolution | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Singh, P.K, Gupta, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2018-07-02 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of the complex of Phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephospho Coenzyme A at 2.74 A resolution
To Be Published
|
|
1A5P
 
 | | C[40,95]A VARIANT OF BOVINE PANCREATIC RIBONUCLEASE A | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Pearson, M.A, Karplus, P.A, Dodge, R.W, Laity, J.H, Scheraga, H.A. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of two mutants that have implications for the folding of bovine pancreatic ribonuclease A.
Protein Sci., 7, 1998
|
|
5FB6
 
 | | Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering | | Descriptor: | Insulin Chain A, Insulin Chain B | | Authors: | Roedig, P, Duman, R, Sanchez-Weatherby, J, Vartiainen, I, Burkhardt, A, Warmer, M, David, C, Wagner, A, Meents, A. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Room-temperature macromolecular crystallography using a micro-patterned silicon chip with minimal background scattering.
J.Appl.Crystallogr., 49, 2016
|
|
4F3C
 
 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with butyl-thio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Haapalainen, A.M, Thomas, K, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Salmonella enterica with butyl-thio-DADMe-Immucillin-A
Structure, 21, 2013
|
|
4RAT
 
 | |
6SEE
 
 | | Class2A : CENP-A nucleosome in complex with CENP-C central region | | Descriptor: | Centromere protein C, DNA (145-MER), Histone H2A type 2-A, ... | | Authors: | Ali-Ahmad, A, Bilokapic, S, Schafer, I.B, Halic, M, Sekulic, N. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CENP-C unwraps the human CENP-A nucleosome through the H2A C-terminal tail.
Embo Rep., 20, 2019
|
|
