1SDY
 
 | | STRUCTURE SOLUTION AND MOLECULAR DYNAMICS REFINEMENT OF THE YEAST CU,ZN ENZYME SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Djinovic, K, Gatti, G, Coda, A, Antolini, L, Pelosi, G, Desideri, A, Falconi, M, Marmocchi, F, Rotilio, G, Bolognesi, M. | | Deposit date: | 1991-06-14 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure solution and molecular dynamics refinement of the yeast Cu,Zn enzyme superoxide dismutase.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
1BAB
 
 | |
1OVB
 
 | | THE MECHANISM OF IRON UPTAKE BY TRANSFERRINS: THE STRUCTURE OF AN 18KD NII-DOMAIN FRAGMENT AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CARBONATE ION, FE (III) ION, OVOTRANSFERRIN | | Authors: | Kuser, P, Lindley, P, Sarra, R. | | Deposit date: | 1992-10-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The mechanism of iron uptake by transferrins: the structure of an 18 kDa NII-domain fragment from duck ovotransferrin at 2.3 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4HTC
 
 | | THE REFINED STRUCTURE OF THE HIRUDIN-THROMBIN COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | Tulinsky, A, Rydel, T.J, Bode, W, Huber, R. | | Deposit date: | 1993-06-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Refined structure of the hirudin-thrombin complex.
J.Mol.Biol., 221, 1991
|
|
1ASL
 
 | |
1ASN
 
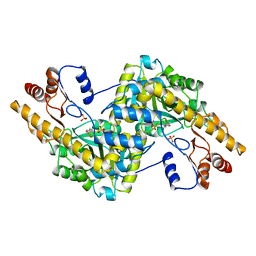 | |
1RAE
 
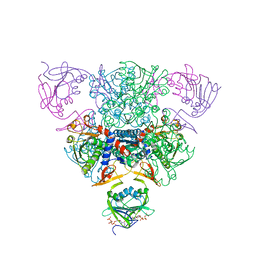 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
2TUN
 
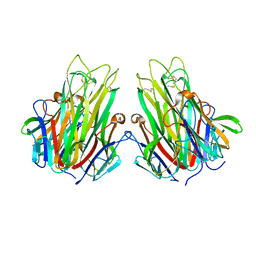 | |
1RAF
 
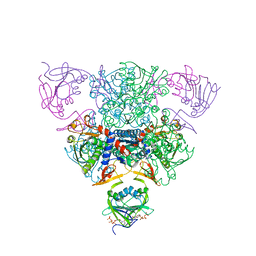 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1PDG
 
 | | CRYSTAL STRUCTURE OF HUMAN PLATELET-DERIVED GROWTH FACTOR BB | | Descriptor: | PLATELET-DERIVED GROWTH FACTOR BB | | Authors: | Oefner, C, Darcy, A.D, Winkler, F.K, Eggimann, B, Hosnag, M. | | Deposit date: | 1992-07-14 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human platelet-derived growth factor BB.
EMBO J., 11, 1992
|
|
2SPL
 
 | | A NOVEL SITE-DIRECTED MUTANT OF MYOGLOBIN WITH AN UNUSUALLY HIGH O2 AFFINITY AND LOW AUTOOXIDATION RATE | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Quillin, M.L, Arduini, R.M, Phillips Jr, G.N. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel site-directed mutant of myoglobin with an unusually high O2 affinity and low autooxidation rate.
J.Biol.Chem., 267, 1992
|
|
1PAF
 
 | | THE 2.5 ANGSTROMS STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Monzingo, A.F, Collins, E.J, Ernst, S.R, Irvin, J.D, Robertus, J.D. | | Deposit date: | 1992-10-19 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 A structure of pokeweed antiviral protein.
J.Mol.Biol., 233, 1993
|
|
6INS
 
 | | X-RAY ANALYSIS OF THE SINGLE CHAIN B29-A1 PEPTIDE-LINKED INSULIN MOLECULE. A COMPLETELY INACTIVE ANALOGUE | | Descriptor: | INSULIN, ZINC ION | | Authors: | Derewenda, U, Derewenda, Z, Dodson, E.J, Dodson, G.G, Bing, X, Markussen, J. | | Deposit date: | 1992-11-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analysis of the single chain B29-A1 peptide-linked insulin molecule. A completely inactive analogue.
J.Mol.Biol., 220, 1991
|
|
1PEK
 
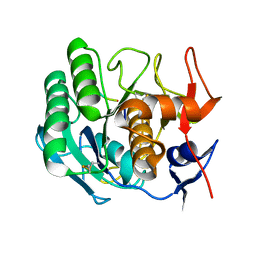 | | STRUCTURE OF THE COMPLEX OF PROTEINASE K WITH A SUBSTRATE-ANALOGUE HEXA-PEPTIDE INHIBITOR AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | D-DAL-ALA-NH2, PEPTIDE PRO-ALA-PRO-PHE, PROTEINASE K | | Authors: | Betzel, C, Singh, T.P, Visanji, M, Peters, K, Fittkau, S, Saenger, W, Wilson, K.S. | | Deposit date: | 1993-01-19 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of proteinase K with a substrate analogue hexapeptide inhibitor at 2.2-A resolution.
J.Biol.Chem., 268, 1993
|
|
1PII
 
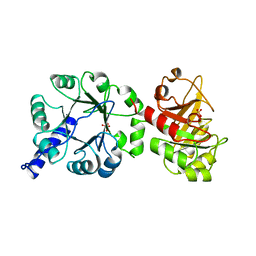 | |
1PER
 
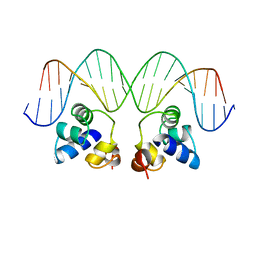 | |
1PAK
 
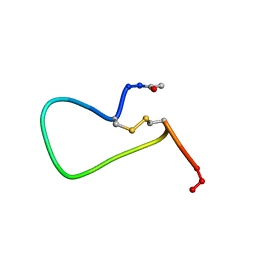 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR, HYDROXIDE ION | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
1BBR
 
 | |
1BAF
 
 | | 2.9 ANGSTROMS RESOLUTION STRUCTURE OF AN ANTI-DINITROPHENYL-SPIN-LABEL MONOCLONAL ANTIBODY FAB FRAGMENT WITH BOUND HAPTEN | | Descriptor: | IGG1-KAPPA AN02 FAB (HEAVY CHAIN), IGG1-KAPPA AN02 FAB (LIGHT CHAIN), N-(2-AMINO-ETHYL)-4,6-DINITRO-N'-(2,2,6,6-TETRAMETHYL-1-OXY-PIPERIDIN-4-YL)-BENZENE-1,3-DIAMINE | | Authors: | Leahy, D.J, Brunger, A.T, Fox, R.O, Hynes, T.R. | | Deposit date: | 1992-01-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 2.9 A resolution structure of an anti-dinitrophenyl-spin-label monoclonal antibody Fab fragment with bound hapten.
J.Mol.Biol., 221, 1991
|
|
1BSE
 
 | |
1PDC
 
 | |
2DRC
 
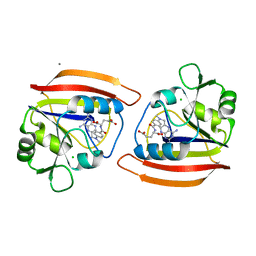 | |
2INT
 
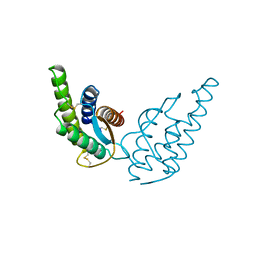 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN INTERLEUKIN-4 | | Descriptor: | INTERLEUKIN-4 | | Authors: | Walter, M.R, Cook, W.J, Zhao, B.G, Cameron Junior, R, Ealick, S.E, Walter Junior, R.L, Reichert, P, Nagabhushan, T.L, Trotta, P.P, Bugg, C.E. | | Deposit date: | 1993-07-22 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of recombinant human interleukin-4.
J.Biol.Chem., 267, 1992
|
|
1PAJ
 
 | | NMR SOLUTION STRUCTURE AND FLEXIBILITY OF A PEPTIDE ANTIGEN REPRESENTING THE RECEPTOR BINDING DOMAIN OF PSEUDOMONAS AERUGINOSA | | Descriptor: | FIMBRIAL PROTEIN PRECURSOR, HYDROXIDE ION | | Authors: | Mcinnes, C, Sonnichsen, F.D, Kay, C.M, Hodges, R.S, Sykes, B.D. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and flexibility of a peptide antigen representing the receptor binding domain of Pseudomonas aeruginosa.
Biochemistry, 32, 1993
|
|
1PIT
 
 | |
